Ready, SET, Go: Post-translational regulation of the histone lysine methylation network in budding yeast
- PMID: 34224729
- PMCID: PMC8329514
- DOI: 10.1016/j.jbc.2021.100939
Ready, SET, Go: Post-translational regulation of the histone lysine methylation network in budding yeast
Abstract
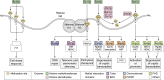
Histone lysine methylation is a key epigenetic modification that regulates eukaryotic transcription. Here, we comprehensively review the function and regulation of the histone methylation network in the budding yeast and model eukaryote, Saccharomyces cerevisiae. First, we outline the lysine methylation sites that are found on histone proteins in yeast (H3K4me1/2/3, H3K36me1/2/3, H3K79me1/2/3, and H4K5/8/12me1) and discuss their biological and cellular roles. Next, we detail the reduced but evolutionarily conserved suite of methyltransferase (Set1p, Set2p, Dot1p, and Set5p) and demethylase (Jhd1p, Jhd2p, Rph1p, and Gis1p) enzymes that are known to control histone lysine methylation in budding yeast cells. Specifically, we illustrate the domain architecture of the methylation enzymes and highlight the structural features that are required for their respective functions and molecular interactions. Finally, we discuss the prevalence of post-translational modifications on yeast histone methylation enzymes and how phosphorylation, acetylation, and ubiquitination in particular are emerging as key regulators of enzyme function. We note that it will be possible to completely connect the histone methylation network to the cell's signaling system, given that all methylation sites and cognate enzymes are known, most phosphosites on the enzymes are known, and the mapping of kinases to phosphosites is tractable owing to the modest set of protein kinases in yeast. Moving forward, we expect that the rich variety of post-translational modifications that decorates the histone methylation machinery will explain many of the unresolved questions surrounding the function and dynamics of this intricate epigenetic network.
Keywords: Saccharomyces cerevisiae; chromatin; demethylase; epigenetics; histone methylation; kinase; methyltransferase; phosphorylation; post-translational modification; post-translational regulation.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





References
-
- Kornberg R.D. Chromatin structure: A repeating unit of histones and DNA. Science. 1974;184:868–871. - PubMed
-
- Murray K. The occurrence of iε-N-methyl lysine in histones. Biochemistry. 1964;3:10–15. - PubMed
-
- Gutierrez R.M., Hnilica L.S. Tissue specificity of histone phosphorylation. Science. 1967;157:1324–1325. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

