Genome-wide association study of stimulant dependence
- PMID: 34226506
- PMCID: PMC8257618
- DOI: 10.1038/s41398-021-01440-5
Genome-wide association study of stimulant dependence
Abstract
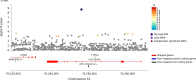
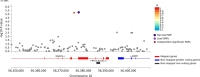
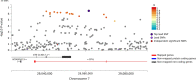
Stimulant dependence is heritable, but specific genetic factors underlying the trait have not been identified. A genome-wide association study for stimulant dependence was performed in a discovery cohort of African- (AA) and European-ancestry (EA) subjects ascertained for genetic studies of alcohol, opioid, and cocaine use disorders. The sample comprised individuals with DSM-IV stimulant dependence (393 EA cases, 5288 EA controls; 155 AA cases, 5603 AA controls). An independent cohort from the family-based Collaborative Study on the Genetics of Alcoholism (532 EA cases, 7635 EA controls; 53 AA cases, AA 3352 controls) was used for replication. One variant in SLC25A16 (rs2394476, p = 3.42 × 10-10, odds ratio [OR] = 3.70) was GWS in AAs. Four other loci showed suggestive evidence, including KCNA4 in AAs (rs11500237, p = 2.99 × 10-7, OR = 2.31) which encodes one of the potassium voltage-gated channel protein that has been linked to several other substance use disorders, and CPVL in the combined population groups (rs1176440, p = 3.05 × 10-7, OR = 1.35), whose expression was previously shown to be upregulated in the prefrontal cortex from users of cocaine, cannabis, and phencyclidine. Analysis of the top GWAS signals revealed a significant enrichment with nicotinic acetylcholine receptor genes (adjusted p = 0.04) and significant pleiotropy between stimulant dependence and alcohol dependence in EAs (padj = 3.6 × 10-3), an anxiety disorder in EAs (padj = 2.1 × 10-4), and ADHD in both AAs (padj = 3.0 × 10-33) and EAs (padj = 6.7 × 10-35). Our results implicate novel genes and pathways as having roles in the etiology of stimulant dependence.
Conflict of interest statement
R.K. is a member of an advisory board for Dicerna Pharmaceuticals and of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which in the past 3 years was sponsored by AbbVie, Alkermes, Amygdala Neurosciences, Arbor Pharmaceuticals, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, and Pfizer. R.K. and J.G. are named as inventors on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018. Other authors do not report any conflicts of interest.
Figures


References
-
- Substance Abuse and Mental Health Services Administration (SAMHSA), D.A.W.N., 2011: National Estimates of Drug-Related Emergency Department Visits. (Substance Abuse and Mental Health Services Administration, 2013).
-
- Hughes, A. et al. Prescription drug use and misuse in the United States: results from the 2015 national survey on drug use and health. NSDUH Data Rev. https://www.samhsa.gov/data/sites/default/files/NSDUH-FFR2-2015/NSDUH-FF... (2016).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

