Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner
- PMID: 34226711
- PMCID: PMC8318886
- DOI: 10.1038/s41564-021-00920-0
Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner
Abstract
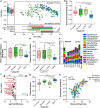
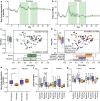
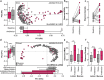
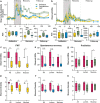
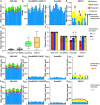
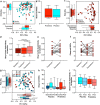
Antimicrobial resistance poses a substantial threat to human health. The gut microbiome is considered a reservoir for potential spread of resistance genes from commensals to pathogens, termed the gut resistome. The impact of probiotics, commonly consumed by many in health or in conjunction with the administration of antibiotics, on the gut resistome is elusive. Reanalysis of gut metagenomes from healthy antibiotics-naïve humans supplemented with an 11-probiotic-strain preparation, allowing direct assessment of the gut resistome in situ along the gastrointestinal (GI) tract, demonstrated that probiotics reduce the number of antibiotic resistance genes exclusively in the gut of colonization-permissive individuals. In mice and in a separate cohort of humans, a course of antibiotics resulted in expansion of the lower GI tract resistome, which was mitigated by autologous faecal microbiome transplantation or during spontaneous recovery. In contrast, probiotics further exacerbated resistome expansion in the GI mucosa by supporting the bloom of strains carrying vancomycin resistance genes but not resistance genes encoded by the probiotic strains. Importantly, the aforementioned effects were not reflected in stool samples, highlighting the importance of direct sampling to analyse the effect of probiotics and antibiotics on the gut resistome. Analysing antibiotic resistance gene content in additional published clinical trials with probiotics further highlighted the importance of person-specific metagenomics-based profiling of the gut resistome using direct sampling. Collectively, these findings suggest opposing person-specific and antibiotic-dependent effects of probiotics on the resistome, whose contribution to the spread of antimicrobial resistance genes along the human GI tract merit further studies.
© 2021. The Author(s).
Conflict of interest statement
E.E. is a consultant to DayTwo and BiomX. None of the topics related to this work involve these or other commercial entities. The remaining authors declare no competing interests.
Figures






Similar articles
-
Good microbes, bad genes? The dissemination of antimicrobial resistance in the human microbiome.Gut Microbes. 2022 Jan-Dec;14(1):2055944. doi: 10.1080/19490976.2022.2055944. Gut Microbes. 2022. PMID: 35332832 Free PMC article. Review.
-
Comparative gut microbiota and resistome profiling of intensive care patients receiving selective digestive tract decontamination and healthy subjects.Microbiome. 2017 Aug 14;5(1):88. doi: 10.1186/s40168-017-0309-z. Microbiome. 2017. PMID: 28803549 Free PMC article.
-
Antibiotic resistomes of healthy pig faecal metagenomes.Microb Genom. 2019 May;5(5):e000272. doi: 10.1099/mgen.0.000272. Epub 2019 May 15. Microb Genom. 2019. PMID: 31091181 Free PMC article.
-
The Human Gut Resistome up to Extreme Longevity.mSphere. 2021 Oct 27;6(5):e0069121. doi: 10.1128/mSphere.00691-21. Epub 2021 Sep 8. mSphere. 2021. PMID: 34494880 Free PMC article.
-
Potential influence of antimicrobial resistance gene content in probiotic bacteria on the gut resistome ecosystems.Front Nutr. 2023 Feb 1;10:1054555. doi: 10.3389/fnut.2023.1054555. eCollection 2023. Front Nutr. 2023. PMID: 36819705 Free PMC article. Review.
Cited by
-
Good microbes, bad genes? The dissemination of antimicrobial resistance in the human microbiome.Gut Microbes. 2022 Jan-Dec;14(1):2055944. doi: 10.1080/19490976.2022.2055944. Gut Microbes. 2022. PMID: 35332832 Free PMC article. Review.
-
Towards routine employment of computational tools for antimicrobial resistance determination via high-throughput sequencing.Brief Bioinform. 2022 Mar 10;23(2):bbac020. doi: 10.1093/bib/bbac020. Brief Bioinform. 2022. PMID: 35212354 Free PMC article.
-
The double-edged sword of probiotic supplementation on gut microbiota structure in Helicobacter pylori management.Gut Microbes. 2022 Jan-Dec;14(1):2108655. doi: 10.1080/19490976.2022.2108655. Gut Microbes. 2022. PMID: 35951774 Free PMC article. Review.
-
Microbiomes in physiology: insights into 21st-century global medical challenges.Exp Physiol. 2022 Apr;107(4):257-264. doi: 10.1113/EP090226. Epub 2022 Feb 14. Exp Physiol. 2022. PMID: 35081663 Free PMC article. Review.
-
Inhibitory effect of some probiotic strains and essential oils on the growth of some foodborne pathogens.Open Vet J. 2024 Jan;14(1):470-480. doi: 10.5455/OVJ.2024.v14.i1.43. Epub 2024 Jan 31. Open Vet J. 2024. PMID: 38633175 Free PMC article.
References
-
- No Time to Wait: Securing the Future from Drug-Resistant Infections. Report to the Secretary-General of the United Nations (Interagency Coordination Group on Antimicrobial Resistance, 2019).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

