A triple threat: the Parastagonospora nodorum SnTox267 effector exploits three distinct host genetic factors to cause disease in wheat
- PMID: 34227112
- PMCID: PMC9292537
- DOI: 10.1111/nph.17601
A triple threat: the Parastagonospora nodorum SnTox267 effector exploits three distinct host genetic factors to cause disease in wheat
Abstract
Parastagonospora nodorum is a fungal pathogen of wheat. As a necrotrophic specialist, it deploys effector proteins that target dominant host susceptibility genes to elicit programmed cell death (PCD). Here we identify and functionally validate the effector targeting the host susceptibility genes Snn2, Snn6 and Snn7. We utilized whole-genome sequencing, association mapping, gene-disrupted mutants, gain-of-function transformants, virulence assays, bioinformatics and quantitative PCR to characterize these interactions. A single proteinaceous effector, SnTox267, targeted Snn2, Snn6 and Snn7 to trigger PCD. Snn2 and Snn6 functioned cooperatively to trigger PCD in a light-dependent pathway, whereas Snn7-mediated PCD functioned in a light-independent pathway. Isolates harboring 20 SnTox267 protein isoforms quantitatively varied in virulence. The diversity and distribution of isoforms varied between populations, indicating adaptation to local selection pressures. SnTox267 deletion resulted in the upregulation of effector genes SnToxA, SnTox1 and SnTox3. We validated a novel effector operating in an inverse-gene-for-gene manner to target three genetically distinct host susceptibility genes and elicit PCD. The discovery of the complementary gene action of Snn2 and Snn6 indicates their potential function in a guard or decoy model. Additionally, differences in light dependency in the elicited pathways and upregulation of unlinked effectors sheds new light onto a complex fungal necrotroph-host interaction.
Keywords: Parastagonospora nodorum; association mapping; fungi; genomics; necrotrophic effector; programmed cell death; wheat.
© No claim to US Government works New Phytologist © 2021 New Phytologist Foundation.
Figures

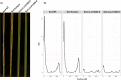
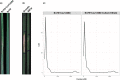

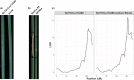



Comment in
-
The rise of necrotrophic effectors.New Phytol. 2022 Jan;233(1):11-14. doi: 10.1111/nph.17811. Epub 2021 Nov 1. New Phytol. 2022. PMID: 34723389 No abstract available.
References
-
- Abeysekara NS, Friesen TL, Keller B, Faris JD. 2009. Identification and characterization of a novel host‐toxin interaction in the wheat‐Stagonospora nodorum pathosystem. Theoretical and Applied Genetics 120: 117–126. - PubMed
-
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nature Biotechnology 37: 420–423. - PubMed
-
- Boller T, Felix G. 2009. A Renaissance of Elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annual Review of Plant Biology 60: 379–406. - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

