Method development and characterisation of the low-molecular-weight peptidome of human wound fluids
- PMID: 34227939
- PMCID: PMC8260221
- DOI: 10.7554/eLife.66876
Method development and characterisation of the low-molecular-weight peptidome of human wound fluids
Abstract
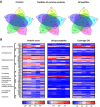
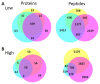
The normal wound healing process is characterised by proteolytic events, whereas infection results in dysfunctional activations by endogenous and bacterial proteases. Peptides, downstream reporters of these proteolytic actions, could therefore serve as a promising tool for diagnosis of wounds. Using mass-spectrometry analyses, we here for the first time characterise the peptidome of human wound fluids. Sterile post-surgical wound fluids were found to contain a high degree of peptides in comparison to human plasma. Analyses of the peptidome from uninfected healing wounds and Staphylococcus aureus -infected wounds identify unique peptide patterns of various proteins, including coagulation and complement factors, proteases, and antiproteinases. Together, the work defines a workflow for analysis of peptides derived from wound fluids and demonstrates a proof-of-concept that such fluids can be used for analysis of qualitative differences of peptide patterns from larger patient cohorts, providing potential biomarkers for wound healing and infection.
Keywords: bacterial infection; biochemistry; biomarkers; chemical biology; human; immunology; inflammation; peptide profiles; peptidomics; plasma; wound fluid.
Plain language summary
Infected wounds and burns represent a serious risk to patients: they can delay healing and, if left untreated, can lead to generalised infection or sepsis, organ failure and death. Wounds and burns get infected when harmful micro-organisms, such as bacteria, enter the wound. Predicting the risk of infections, and detecting them early, could reduce their impact and make treating them easier. A way to distinguish between healing and infected wounds is to study how proteins are broken down in each situation. Proteases are the enzymes that break down proteins, and they are different in healing wounds and infected wounds that are failing to heal. This is because, while the body produces proteases, the bacteria that cause infection do so too. Each protease breaks down proteins in a specific way, resulting in a different set of protein fragments, known as peptides. Together, all the peptides in a wound are referred to as the wound’s ‘peptidome’. Studying the peptidome of a wound could show whether it is infected, and even what type of bacteria might be responsible, which could help identify suitable treatments. Van der Plas et al. used a technique called mass spectrometry to study the peptidome of wounds after surgery. Sterile post-surgical wounds showed high levels of peptides compared to plasma, the liquid component of blood, with up to 4,300 different peptides. Comparing healing wounds to ones infected with the bacterium Staphylococcus aureus revealed that infected wounds contained peptides from about 150 proteins not found in uninfected wounds, while peptides from 90 proteins were unique to uninfected wounds. The peptides exclusive to uninfected wounds included some linked to antimicrobial activity and immune system activity. Van der Plas et al.’s results suggest that analysing the peptidome may be an approach to tracking the healing status of wounds, making it easier to detect infection before symptoms are apparent. The next step will be to study more wounds and identify the reliable peptide markers to use them for diagnostic tests.
© 2021, van der Plas et al.
Conflict of interest statement
Mv, JC, JP, KS, SK, AS No competing interests declared
Figures







References
-
- Abdul S, Dekkers DHW, Ariëns RAS, Leebeek FWG, Rijken DC, Uitte de Willige S. On the localization of the cleavage site in human alpha-2-antiplasmin, involved in the generation of the non-plasminogen binding form. Journal of Thrombosis and Haemostasis. 2020;18:1162–1170. doi: 10.1111/jth.14761. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

