Deep Learning Applied to Automated Segmentation of Geographic Atrophy in Fundus Autofluorescence Images
- PMID: 34228106
- PMCID: PMC8267211
- DOI: 10.1167/tvst.10.8.2
Deep Learning Applied to Automated Segmentation of Geographic Atrophy in Fundus Autofluorescence Images
Abstract
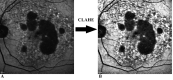
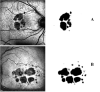
Purpose: This study describes the development of a deep learning algorithm based on the U-Net architecture for automated segmentation of geographic atrophy (GA) lesions in fundus autofluorescence (FAF) images.
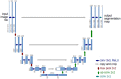
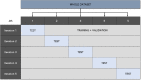
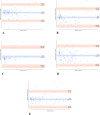
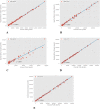
Methods: Image preprocessing and normalization by modified adaptive histogram equalization were used for image standardization to improve effectiveness of deep learning. A U-Net-based deep learning algorithm was developed and trained and tested by fivefold cross-validation using FAF images from clinical datasets. The following metrics were used for evaluating the performance for lesion segmentation in GA: dice similarity coefficient (DSC), DSC loss, sensitivity, specificity, mean absolute error (MAE), accuracy, recall, and precision.
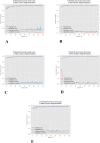
Results: In total, 702 FAF images from 51 patients were analyzed. After fivefold cross-validation for lesion segmentation, the average training and validation scores were found for the most important metric, DSC (0.9874 and 0.9779), for accuracy (0.9912 and 0.9815), for sensitivity (0.9955 and 0.9928), and for specificity (0.8686 and 0.7261). Scores for testing were all similar to the validation scores. The algorithm segmented GA lesions six times more quickly than human performance.
Conclusions: The deep learning algorithm can be implemented using clinical data with a very high level of performance for lesion segmentation. Automation of diagnostics for GA assessment has the potential to provide savings with respect to patient visit duration, operational cost and measurement reliability in routine GA assessments.
Translational relevance: A deep learning algorithm based on the U-Net architecture and image preprocessing appears to be suitable for automated segmentation of GA lesions on clinical data, producing fast and accurate results.
Conflict of interest statement
Disclosure:
Figures


References
-
- Wong WL, Su X, Li X, et al. .. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Global Health. 2014; 2: e106–e116. - PubMed
-
- Holz FG, Strauss EC, Schmitz-Valckenberg S, van Lookeren Campagne M.. Geographic atrophy: clinical features and potential therapeutic approaches. Ophthalmology. 2014; 121: 1079–1091. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

