Relative synonymous codon usage of ORF1ab in SARS-CoV-2 and SARS-CoV
- PMID: 34228320
- PMCID: PMC8258482
- DOI: 10.1007/s13258-021-01136-6
Relative synonymous codon usage of ORF1ab in SARS-CoV-2 and SARS-CoV
Abstract
Background: COVID-19, as a novel coronavirus disease caused by new coronavirus SARS-CoV-2, spreads all over the world, and brings harm to human in many countries. Humans suffered a lot from both SARS-CoV-2 now and by SARS-CoV in the year 2003. It is important to understand the differences and the relationships between these two types of viruses.
Objective: To compare relative synonymous codon usage of ORF1ab gene in SARS-CoV-2 and SARS-CoV, relative synonymous codon usage of their genomes are studied in this paper from the bioinformatics perspective.
Methods: The ORF1ab gene, which is an important non-structural polyprotein coding gene and now used for nucleic acid detection markers in many measurement method, in both SARS-CoV-2 (30 strains) and SARS-CoV (20 strains) are considered to be the research object in the present paper. The relative synonymous codon usage values of the ORF1ab gene are calculated to characterize the differences and the evolutionary characteristics among 50 strains.
Results: There is a significant difference between SARS-CoV and SARS-CoV-2 when the relative synonymous codon usage value of ORF1ab genes is concerned. The results suggest that codon usage pattern of SARS-CoV is more similar to human than that of the SARS-CoV-2, and that the inner difference in SARS-CoV-2 strains is larger than that of SARS-CoV, which denote the larger diversity exits in the SARS-CoV-2 virus.
Conclusion: These results show that the relative synonymous codon usage values in the coronavirus could be used for further research on their evolutionary phenomenon.
Keywords: COVID-19; Codon usage pattern; Coronavirus; Gene evolution; Relative synonymous codon usage; SARS-CoV; SARS-CoV-2.
© 2021. The Genetics Society of Korea.
Conflict of interest statement
None.
Figures

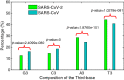
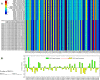

Similar articles
-
Analysis of SARS-CoV-2 synonymous codon usage evolution throughout the COVID-19 pandemic.Virology. 2022 Mar;568:56-71. doi: 10.1016/j.virol.2022.01.011. Epub 2022 Feb 2. Virology. 2022. PMID: 35134624 Free PMC article.
-
Codon usage divergence in Delta variants (B.1.617.2) of SARS-CoV-2.Infect Genet Evol. 2022 Jan;97:105175. doi: 10.1016/j.meegid.2021.105175. Epub 2021 Dec 3. Infect Genet Evol. 2022. PMID: 34871776 Free PMC article.
-
Characterization of accessory genes in coronavirus genomes.Virol J. 2020 Aug 27;17(1):131. doi: 10.1186/s12985-020-01402-1. Virol J. 2020. PMID: 32854725 Free PMC article.
-
Unpacking Pandora From Its Box: Deciphering the Molecular Basis of the SARS-CoV-2 Coronavirus.Int J Mol Sci. 2020 Dec 31;22(1):386. doi: 10.3390/ijms22010386. Int J Mol Sci. 2020. PMID: 33396557 Free PMC article. Review.
-
A nidovirus perspective on SARS-CoV-2.Biochem Biophys Res Commun. 2021 Jan 29;538:24-34. doi: 10.1016/j.bbrc.2020.11.015. Epub 2020 Nov 13. Biochem Biophys Res Commun. 2021. PMID: 33413979 Free PMC article. Review.
Cited by
-
Componential usage patterns in dengue 4 viruses reveal their better evolutionary adaptation to humans.Front Microbiol. 2022 Sep 20;13:935678. doi: 10.3389/fmicb.2022.935678. eCollection 2022. Front Microbiol. 2022. PMID: 36204606 Free PMC article.
-
Analysis of SARS-CoV-2 synonymous codon usage evolution throughout the COVID-19 pandemic.Virology. 2022 Mar;568:56-71. doi: 10.1016/j.virol.2022.01.011. Epub 2022 Feb 2. Virology. 2022. PMID: 35134624 Free PMC article.
-
Intraspecific and interspecific variations in the synonymous codon usage in mitochondrial genomes of 8 pleurotus strains.BMC Genomics. 2024 May 10;25(1):456. doi: 10.1186/s12864-024-10374-3. BMC Genomics. 2024. PMID: 38730418 Free PMC article.
-
Molecular characterization of virulent genes in Pseudomonas aeruginosa based on componential usage divergence.Sci Rep. 2025 Apr 2;15(1):11246. doi: 10.1038/s41598-025-95579-6. Sci Rep. 2025. PMID: 40175567 Free PMC article.
References
-
- Bartolini B, Rueca M, Gruber CEM, Messina F, Carletti F, Giombini E, Lalle E, Bordi L, Matusali G, Colavita F, Castilletti C, Vairo F, Ippolito G, Capobianchi MR, Di Caro A. SARS-CoV-2 phylogenetic analysis, Lazio Region, Italy, February–March 2020. Emerg Infect Dis. 2020;26(8):1842–1845. doi: 10.3201/eid2608.201525. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
