Aging-related upregulation of the homeobox gene caudal represses intestinal stem cell differentiation in Drosophila
- PMID: 34228720
- PMCID: PMC8284806
- DOI: 10.1371/journal.pgen.1009649
Aging-related upregulation of the homeobox gene caudal represses intestinal stem cell differentiation in Drosophila
Erratum in
-
Correction: Aging-related upregulation of the homeobox gene caudal represses intestinal stem cell differentiation in Drosophila.PLoS Genet. 2025 Sep 4;21(9):e1011846. doi: 10.1371/journal.pgen.1011846. eCollection 2025 Sep. PLoS Genet. 2025. PMID: 40906680
Abstract
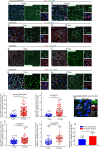
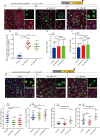
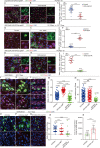
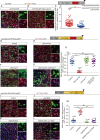
The differentiation efficiency of adult stem cells undergoes a significant decline in aged animals, which is closely related to the decline in organ function and age-associated diseases. However, the underlying mechanisms that ultimately lead to this observed decline of the differentiation efficiency of stem cells remain largely unclear. This study investigated Drosophila midguts and identified an obvious upregulation of caudal (cad), which encodes a homeobox transcription factor. This factor is traditionally known as a central regulator of embryonic anterior-posterior body axis patterning. This study reports that depletion of cad in intestinal stem/progenitor cells promotes quiescent intestinal stem cells (ISCs) to become activate and produce enterocytes in the midgut under normal gut homeostasis conditions. However, overexpression of cad results in the failure of ISC differentiation and intestinal epithelial regeneration after injury. Moreover, this study suggests that cad prevents intestinal stem/progenitor cell differentiation by modulating the Janus kinase/signal transducers and activators of the transcription pathway and Sox21a-GATAe signaling cascade. Importantly, the reduction of cad expression in intestinal stem/progenitor cells restrained age-associated gut hyperplasia in Drosophila. This study identified a function of the homeobox gene cad in the modulation of adult stem cell differentiation and suggested a potential gene target for the treatment of age-related diseases induced by age-related stem cell dysfunction.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
The role of p38b MAPK in age-related modulation of intestinal stem cell proliferation and differentiation in Drosophila.Aging (Albany NY). 2009 May 21;1(7):637-51. doi: 10.18632/aging.100054. Aging (Albany NY). 2009. PMID: 20157545 Free PMC article.
-
Non-autonomous crosstalk between the Jak/Stat and Egfr pathways mediates Apc1-driven intestinal stem cell hyperplasia in the Drosophila adult midgut.Development. 2012 Dec;139(24):4524-35. doi: 10.1242/dev.078261. Development. 2012. PMID: 23172913
-
Paracrine unpaired signaling through the JAK/STAT pathway controls self-renewal and lineage differentiation of drosophila intestinal stem cells.J Mol Cell Biol. 2010 Feb;2(1):37-49. doi: 10.1093/jmcb/mjp028. Epub 2009 Sep 30. J Mol Cell Biol. 2010. PMID: 19797317
-
Intestinal stem cell response to injury: lessons from Drosophila.Cell Mol Life Sci. 2016 Sep;73(17):3337-49. doi: 10.1007/s00018-016-2235-9. Epub 2016 May 2. Cell Mol Life Sci. 2016. PMID: 27137186 Free PMC article. Review.
-
Intestinal stem cells in the adult Drosophila midgut.Exp Cell Res. 2011 Nov 15;317(19):2780-8. doi: 10.1016/j.yexcr.2011.07.020. Epub 2011 Aug 11. Exp Cell Res. 2011. PMID: 21856297 Free PMC article. Review.
Cited by
-
Stress Induced Activation of LTR Retrotransposons in the Drosophila melanogaster Genome.Life (Basel). 2023 Nov 28;13(12):2272. doi: 10.3390/life13122272. Life (Basel). 2023. PMID: 38137873 Free PMC article.
-
Colonic Aging and Colorectal Cancer: An Unignorable Interplay and Its Translational Implications.Biology (Basel). 2025 Jul 3;14(7):805. doi: 10.3390/biology14070805. Biology (Basel). 2025. PMID: 40723364 Free PMC article. Review.
-
Drosophila AHR limits tumor growth and stem cell proliferation in the intestine.Wellcome Open Res. 2025 Apr 25;10:38. doi: 10.12688/wellcomeopenres.23515.3. eCollection 2025. Wellcome Open Res. 2025. PMID: 40212817 Free PMC article.
-
Antioxidant Effects of Caffeic Acid Lead to Protection of Drosophila Intestinal Stem Cell Aging.Front Cell Dev Biol. 2021 Sep 9;9:735483. doi: 10.3389/fcell.2021.735483. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34568344 Free PMC article.
-
De novo apical domain formation inside the Drosophila adult midgut epithelium.Elife. 2022 Sep 28;11:e76366. doi: 10.7554/eLife.76366. Elife. 2022. PMID: 36169289 Free PMC article.
References
-
- Ambrosi TH, Scialdone A, Graja A, Gohlke S, Jank AM, Bocian C, et al. Adipocyte Accumulation in the Bone Marrow during Obesity and Aging Impairs Stem Cell-Based Hematopoietic and Bone Regeneration. Cell Stem Cell. 2017;20(6):771–84 e6. Epub 2017/03/24. doi: 10.1016/j.stem.2017.02.009 ; PubMed Central PMCID: PMC5459794. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

