Label-retention expansion microscopy
- PMID: 34228783
- PMCID: PMC8266563
- DOI: 10.1083/jcb.202105067
Label-retention expansion microscopy
Abstract
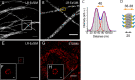
Expansion microscopy (ExM) increases the effective resolving power of any microscope by expanding the sample with swellable hydrogel. Since its invention, ExM has been successfully applied to a wide range of cell, tissue, and animal samples. Still, fluorescence signal loss during polymerization and digestion limits molecular-scale imaging using ExM. Here, we report the development of label-retention ExM (LR-ExM) with a set of trifunctional anchors that not only prevent signal loss but also enable high-efficiency labeling using SNAP and CLIP tags. We have demonstrated multicolor LR-ExM for a variety of subcellular structures. Combining LR-ExM with superresolution stochastic optical reconstruction microscopy (STORM), we have achieved molecular resolution in the visualization of polyhedral lattice of clathrin-coated pits in situ.
© 2021 Shi et al.
Figures