Consensus clustering applied to multi-omics disease subtyping
- PMID: 34229612
- PMCID: PMC8259015
- DOI: 10.1186/s12859-021-04279-1
Consensus clustering applied to multi-omics disease subtyping
Abstract
Background: Facing the diversity of omics data and the difficulty of selecting one result over all those produced by several methods, consensus strategies have the potential to reconcile multiple inputs and to produce robust results.
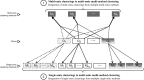
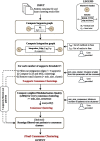
Results: Here, we introduce ClustOmics, a generic consensus clustering tool that we use in the context of cancer subtyping. ClustOmics relies on a non-relational graph database, which allows for the simultaneous integration of both multiple omics data and results from various clustering methods. This new tool conciliates input clusterings, regardless of their origin, their number, their size or their shape. ClustOmics implements an intuitive and flexible strategy, based upon the idea of evidence accumulation clustering. ClustOmics computes co-occurrences of pairs of samples in input clusters and uses this score as a similarity measure to reorganize data into consensus clusters.
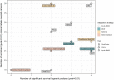
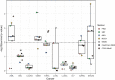
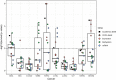
Conclusion: We applied ClustOmics to multi-omics disease subtyping on real TCGA cancer data from ten different cancer types. We showed that ClustOmics is robust to heterogeneous qualities of input partitions, smoothing and reconciling preliminary predictions into high-quality consensus clusters, both from a computational and a biological point of view. The comparison to a state-of-the-art consensus-based integration tool, COCA, further corroborated this statement. However, the main interest of ClustOmics is not to compete with other tools, but rather to make profit from their various predictions when no gold-standard metric is available to assess their significance.
Availability: The ClustOmics source code, released under MIT license, and the results obtained on TCGA cancer data are available on GitHub: https://github.com/galadrielbriere/ClustOmics .
Keywords: Consensus clustering; Data integration; Disease subtyping; Multi-omic data.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
NEMO: cancer subtyping by integration of partial multi-omic data.Bioinformatics. 2019 Sep 15;35(18):3348-3356. doi: 10.1093/bioinformatics/btz058. Bioinformatics. 2019. PMID: 30698637 Free PMC article.
-
CSSEC: An adaptive approach integrating consensus and specific self-expressive coefficients for multi-omics cancer subtyping.Methods. 2025 Mar;235:26-33. doi: 10.1016/j.ymeth.2025.01.016. Epub 2025 Jan 27. Methods. 2025. PMID: 39880224
-
Supervised Graph Clustering for Cancer Subtyping Based on Survival Analysis and Integration of Multi-Omic Tumor Data.IEEE/ACM Trans Comput Biol Bioinform. 2022 Mar-Apr;19(2):1193-1202. doi: 10.1109/TCBB.2020.3010509. Epub 2022 Apr 1. IEEE/ACM Trans Comput Biol Bioinform. 2022. PMID: 32750893
-
Multi-omic and multi-view clustering algorithms: review and cancer benchmark.Nucleic Acids Res. 2018 Nov 16;46(20):10546-10562. doi: 10.1093/nar/gky889. Nucleic Acids Res. 2018. PMID: 30295871 Free PMC article. Review.
-
IOAT: an interactive tool for statistical analysis of omics data and clinical data.BMC Bioinformatics. 2021 Jun 15;22(1):326. doi: 10.1186/s12859-021-04253-x. BMC Bioinformatics. 2021. PMID: 34130622 Free PMC article. Review.
Cited by
-
Artificial intelligence in ovarian cancer drug resistance advanced 3PM approach: subtype classification and prognostic modeling.EPMA J. 2024 Jul 13;15(3):525-544. doi: 10.1007/s13167-024-00374-4. eCollection 2024 Sep. EPMA J. 2024. PMID: 39239109
-
Identification and validation of a prognostic signature of cuproptosis-related genes for esophageal squamous cell carcinoma.Aging (Albany NY). 2023 Sep 2;15(17):8993-9021. doi: 10.18632/aging.205012. Epub 2023 Sep 2. Aging (Albany NY). 2023. PMID: 37665670 Free PMC article.
-
Integrated bioinformatic analysis of mitochondrial metabolism-related genes in acute myeloid leukemia.Front Immunol. 2023 Apr 17;14:1120670. doi: 10.3389/fimmu.2023.1120670. eCollection 2023. Front Immunol. 2023. PMID: 37138869 Free PMC article.
-
A Novel Neutrophil Extracellular Trap Signature Predicts Patient Chemotherapy Resistance and Prognosis in Lung Adenocarcinoma.Mol Biotechnol. 2025 May;67(5):1939-1957. doi: 10.1007/s12033-024-01170-1. Epub 2024 May 11. Mol Biotechnol. 2025. PMID: 38734842
-
Three decades of advancements in osteoarthritis research: insights from transcriptomic, proteomic, and metabolomic studies.Osteoarthritis Cartilage. 2024 Apr;32(4):385-397. doi: 10.1016/j.joca.2023.11.019. Epub 2023 Dec 2. Osteoarthritis Cartilage. 2024. PMID: 38049029 Free PMC article. Review.
References
-
- Wang H, Nie F, Huang H. Multi-view clustering and feature learning via structured sparsity. In: Proceedings of the 30th international conference on international conference on machine learning—volume 28. ICML’13, pp. 352–360. JMLR.org, Atlanta, GA, USA. 2013.
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

