Long-chain polyphosphates impair SARS-CoV-2 infection and replication
- PMID: 34230209
- PMCID: PMC8432949
- DOI: 10.1126/scisignal.abe5040
Long-chain polyphosphates impair SARS-CoV-2 infection and replication
Abstract
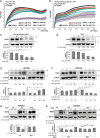
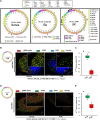
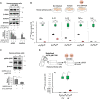
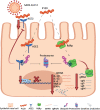
Inorganic polyphosphates (polyPs) are linear polymers composed of repeated phosphate (PO4 3-) units linked together by multiple high-energy phosphoanhydride bonds. In addition to being a source of energy, polyPs have cytoprotective and antiviral activities. Here, we investigated the antiviral activities of long-chain polyPs against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. In molecular docking analyses, polyPs interacted with several conserved amino acid residues in angiotensin-converting enzyme 2 (ACE2), the host receptor that facilitates virus entry, and in viral RNA-dependent RNA polymerase (RdRp). ELISA and limited proteolysis assays using nano- LC-MS/MS mapped polyP120 binding to ACE2, and site-directed mutagenesis confirmed interactions between ACE2 and SARS-CoV-2 RdRp and identified the specific amino acid residues involved. PolyP120 enhanced the proteasomal degradation of both ACE2 and RdRp, thus impairing replication of the British B.1.1.7 SARS-CoV-2 variant. We thus tested polyPs for functional interactions with the virus in SARS-CoV-2-infected Vero E6 and Caco2 cells and in primary human nasal epithelial cells. Delivery of a nebulized form of polyP120 reduced the amounts of viral positive-sense genomic and subgenomic RNAs, of RNA transcripts encoding proinflammatory cytokines, and of viral structural proteins, thereby presenting SARS-CoV-2 infection in cells in vitro.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures


References
-
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F. G., Horby P. W., Cao B., Wang C., Remdesivir in adults with severe COVID-19: A randomised, double-blind, placebo-controlled, multicentre trial. Lancet 395, 1569–1578 (2020). - PMC - PubMed
-
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). - PMC - PubMed
-
- Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., Liu L., Shan H., Lei C.-l., Hui D. S. C., Du B., Li L.-j., Zeng G., Yuen K.-Y., Chen R.-c., Tang C.-l., Wang T., Chen P.-y., Xiang J., Li S.-y., Wang J.-l., Liang Z.-j., Peng Y.-x., Wei L., Liu Y., Hu Y.-h., Peng P., Wang J.-m., Liu J.-y., Chen Z., Li G., Zheng Z.-j., Qiu S.-q., Luo J., Ye C.-j., Zhu S.-y., Zhong N.-s.; China Medical Treatment Expert Group for Covid-19 , Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 382, 1708–1720 (2020). - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

