A bacteriocin-based treatment option for Staphylococcus haemolyticus biofilms
- PMID: 34230527
- PMCID: PMC8260761
- DOI: 10.1038/s41598-021-93158-z
A bacteriocin-based treatment option for Staphylococcus haemolyticus biofilms
Abstract
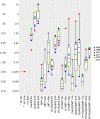
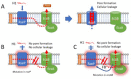
Bacteriocins are ribosomally-synthesized antimicrobial peptides, showing great potential as novel treatment options for multidrug-resistant pathogens. In this study, we designed a novel hybrid bacteriocin, Hybrid 1 (H1), by combing the N-terminal part and the C-terminal part of the related bacteriocins enterocin K1 (K1) and enterocin EJ97 (EJ97), respectively. Like the parental bacteriocins, H1 used the membrane-bound protease RseP as receptor, however, it differed from the others in the inhibition spectrum. Most notably, H1 showed a superior antimicrobial effect towards Staphylococcus haemolyticus-an important nosocomial pathogen. To avoid strain-dependency, we further evaluated H1 against 27 clinical and commensal S. haemolyticus strains, with H1 indeed showing high activity towards all strains. To curtail the rise of resistant mutants and further explore the potential of H1 as a therapeutic agent, we designed a bacteriocin-based formulation where H1 was used in combination with the broad-spectrum bacteriocins micrococcin P1 and garvicin KS. Unlike the individual bacteriocins, the three-component combination was highly effective against planktonic cells and completely eradicated biofilm-associated S. haemolyticus cells in vitro. Most importantly, the formulation efficiently prevented development of resistant mutants as well. These findings indicate the potential of a bacteriocins-based formulation as a treatment option for S. haemolyticus.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

