Identification of potential core genes in esophageal carcinoma using bioinformatics analysis
- PMID: 34232175
- PMCID: PMC8270608
- DOI: 10.1097/MD.0000000000026428
Identification of potential core genes in esophageal carcinoma using bioinformatics analysis
Abstract
Background: Esophageal squamous cell carcinoma (ESCC) is a common human malignancy worldwide. The tumorigenesis mechanism in ESCC is unclear.
Materials and methods: To explore potential therapeutic targets for ESCC, we analyzed 3 microarray datasets (GSE20347, GSE38129, and GSE67269) derived from the gene expression omnibus (GEO) database. Then, the GEO2R tool was used to screen out differently expressed genes (DEGs) between ESCC and normal tissue. Gene ontology function and kyoto encyclopedia of genes and genomes pathway enrichment analysis were performed using the database for annotation, visualization and integrated discovery to identify the pathways and functional annotation of DEGs. Protein-protein interaction of these DEGs was analyzed based on the search tool for the retrieval of interacting genes database and visualized by Cytoscape software. In addition, we used encyclopedia of RNA interactomes (ENCORI), gene expression profiling interactive analysis (GEPIA), and the human protein atlas to confirm the expression of hub genes in ESCC. Finally, GEPIA was used to evaluate the prognostic value of hub genes expression in ESCC patients and we estimated the associations between hub genes expression and immune cell populations (B Cell, CD8+ T Cell, CD4+ T Cell, Macrophage, Neutrophil, and Dendritic Cell) in esophageal carcinoma (ESCA) using tumor immune estimation resource (TIMER).
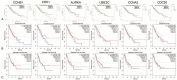
Results: In this study, 707 DEGs (including 385 upregulated genes and 322 downregulated genes) and 6 hub genes (cyclin B1 [CCNB1], cyclin dependent kinase 1 [CDK1], aurora kinase A [AURKA], ubiquitin conjugating enzyme E2C [UBE2C], cyclin A2 [CCNA2], and cell division cycle 20 [CDC20]) were identified. All of the 6 hub genes were highly expressed in ESCC tissues. Among of them, only CCNB1 and CDC20 were associated with stage of ESCC and all of them were not associated with survival time of patients.
Conclusion: DEGs and hub genes were confirmed in our study, providing a thorough, scientific and comprehensive research goals for the pathogenesis of ESCC.
Copyright © 2021 the Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
The authors have no conflicts of interests to disclose.
Figures





References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA 2018;68:394–424. - PubMed
MeSH terms
Substances
Grants and funding
- 2018YZ001-1/Fujian Province Major Science and Technology Program
- 2017XQ1069/Startup Fund for scientific research, Fujian Medical University
- 2017XQ1032/Startup Fund for scientific research, Fujian Medical University
- 2018Y9034/Joint Funds for the Innovation of Science and Technology, Fujian Province
- 2019QH1023/Startup Fund for scientific research, Fujian Medical University
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

