SUMO paralogue-specific functions revealed through systematic analysis of human knockout cell lines and gene expression data
- PMID: 34232706
- PMCID: PMC8684707
- DOI: 10.1091/mbc.E21-01-0031
SUMO paralogue-specific functions revealed through systematic analysis of human knockout cell lines and gene expression data
Abstract
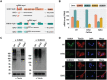
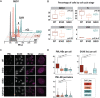
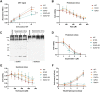
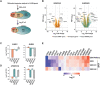
The small ubiquitin-related modifiers (SUMOs) regulate nearly every aspect of cellular function, from gene expression in the nucleus to ion transport at the plasma membrane. In humans, the SUMO pathway has five SUMO paralogues with sequence homologies that range from 45% to 97%. SUMO1 and SUMO2 are the most distantly related paralogues and also the best studied. To what extent SUMO1, SUMO2, and the other paralogues impart unique and nonredundant effects on cellular functions, however, has not been systematically examined and is therefore not fully understood. For instance, knockout studies in mice have revealed conflicting requirements for the paralogues during development and studies in cell culture have relied largely on transient paralogue overexpression or knockdown. To address the existing gap in understanding, we first analyzed SUMO paralogue gene expression levels in normal human tissues and found unique patterns of SUMO1-3 expression across 30 tissue types, suggesting paralogue-specific functions in adult human tissues. To systematically identify and characterize unique and nonredundant functions of the SUMO paralogues in human cells, we next used CRISPR-Cas9 to knock out SUMO1 and SUMO2 expression in osteosarcoma (U2OS) cells. Analysis of these knockout cell lines revealed essential functions for SUMO1 and SUMO2 in regulating cellular morphology, promyelocytic leukemia (PML) nuclear body structure, responses to proteotoxic and genotoxic stress, and control of gene expression. Collectively, our findings reveal nonredundant regulatory roles for SUMO1 and SUMO2 in controlling essential cellular processes and provide a basis for more precise SUMO-targeting therapies.
Figures




References
-
- Adorisio S, Fierabracci A, Muscari I, Liberati AM, Ayroldi E, Migliorati G, Thuy TT, Riccardi C, Delfino DV (2017). SUMO proteins: guardians of immune system. J Autoimmun 84, 21–28. - PubMed
-
- Alkuraya FS, Saadi I, Lund JJ, Turbe-Doan A, Morton CC, Maas RL (2006). SUMO1 haploinsufficiency leads to cleft lip and palate. Science 313, 1751. - PubMed
-
- Augustine RC, Vierstra RD (2018). SUMOylation: re-wiring the plant nucleus during stress and development. Curr Opin Plant Biol 45, 143–154. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

