A homozygous stop-gain variant in ARHGAP42 is associated with childhood interstitial lung disease, systemic hypertension, and immunological findings
- PMID: 34232960
- PMCID: PMC8289122
- DOI: 10.1371/journal.pgen.1009639
A homozygous stop-gain variant in ARHGAP42 is associated with childhood interstitial lung disease, systemic hypertension, and immunological findings
Abstract
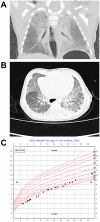
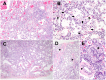
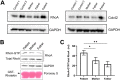
ARHGAP42 encodes Rho GTPase activating protein 42 that belongs to a member of the GTPase Regulator Associated with Focal Adhesion Kinase (GRAF) family. ARHGAP42 is involved in blood pressure control by regulating vascular tone. Despite these findings, disorders of human variants in the coding part of ARHGAP42 have not been reported. Here, we describe an 8-year-old girl with childhood interstitial lung disease (chILD), systemic hypertension, and immunological findings who carries a homozygous stop-gain variant (c.469G>T, p.(Glu157Ter)) in the ARHGAP42 gene. The family history is notable for both parents with hypertension. Histopathological examination of the proband lung biopsy showed increased mural smooth muscle in small airways and alveolar septa, and concentric medial hypertrophy in pulmonary arteries. ARHGAP42 stop-gain variant in the proband leads to exon 5 skipping, and reduced ARHGAP42 levels, which was associated with enhanced RhoA and Cdc42 expression. This is the first report linking a homozygous stop-gain variant in ARHGAP42 with a chILD disorder, systemic hypertension, and immunological findings in human patient. Evidence of smooth muscle hypertrophy on lung biopsy and an increase in RhoA/ROCK signaling in patient cells suggests the potential mechanistic link between ARHGAP42 deficiency and the development of chILD disorder.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Bai X, Dee R, Mangum KD, Mack CP, Taylor JM. RhoA signaling and blood pressure: The consequence of failing to “Tone it Down”. World Journal of Hypertension. 2016;6(1):18–35.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

