Glioma stem cell-derived exosomal miR-944 reduces glioma growth and angiogenesis by inhibiting AKT/ERK signaling
- PMID: 34233294
- PMCID: PMC8386563
- DOI: 10.18632/aging.203243
Glioma stem cell-derived exosomal miR-944 reduces glioma growth and angiogenesis by inhibiting AKT/ERK signaling
Abstract
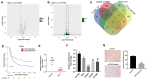
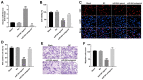
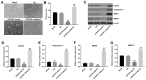
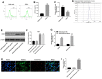
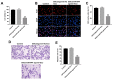
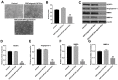
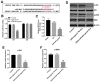
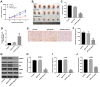
In this study, we investigated the regulatory role of exosomal microRNA-944 (miR-944) derived from glioma stem cells (GSCs) in glioma progression and angiogenesis. Bioinformatics analysis showed that miR-944 levels were significantly lower in high-grade gliomas (HGGs) than low-grade gliomas in the Chinese Glioma Genome Atlas and The Cancer Genome Atlas datasets. The overall survival rates were significantly shorter for glioma patients expressing low miR-944 levels than high miR-944 levels. GSC-derived exosomal miR-944 significantly decreased in vitro proliferation, migration, and tube formation by human umbilical vein endothelial cells (HUVECs). Targetscan and dual luciferase reporter assays demonstrated that miR-944 directly targets the 3'UTR of VEGFC. In vivo mouse studies demonstrated that injection of agomiR-944 directly into tumors 3 weeks after xenografting glioma cells significantly reduced tumor growth and angiogenesis. GSC-derived exosomal miR-944 significantly reduced VEGFC levels and suppressed activation of AKT/ERK signaling pathways in HUVECs and xenograft glioma cell tumors. These findings demonstrate that GSC-derived exosomal miR-944 inhibits glioma growth, progression, and angiogenesis by suppressing VEGFC expression and inhibiting the AKT/ERK signaling pathway.
Keywords: VEGFC; angiogenesis; exosome; glioma stem cells; microRNA-944.
Conflict of interest statement
Figures
Similar articles
-
MicroRNA-30a suppresses self-renewal and tumorigenicity of glioma stem cells by blocking the NT5E-dependent Akt signaling pathway.FASEB J. 2020 Apr;34(4):5128-5143. doi: 10.1096/fj.201802629RR. Epub 2020 Feb 17. FASEB J. 2020. PMID: 32067282
-
Glioma stem cells-derived exosomal miR-26a promotes angiogenesis of microvessel endothelial cells in glioma.J Exp Clin Cancer Res. 2019 May 17;38(1):201. doi: 10.1186/s13046-019-1181-4. J Exp Clin Cancer Res. 2019. PMID: 31101062 Free PMC article.
-
The U2AF2 /circRNA ARF1/miR-342-3p/ISL2 feedback loop regulates angiogenesis in glioma stem cells.J Exp Clin Cancer Res. 2020 Sep 7;39(1):182. doi: 10.1186/s13046-020-01691-y. J Exp Clin Cancer Res. 2020. PMID: 32894165 Free PMC article.
-
Current Understanding of Exosomal MicroRNAs in Glioma Immune Regulation and Therapeutic Responses.Front Immunol. 2022 Jan 14;12:813747. doi: 10.3389/fimmu.2021.813747. eCollection 2021. Front Immunol. 2022. PMID: 35095909 Free PMC article. Review.
-
MCPIP1 promotes cell proliferation, migration and angiogenesis of glioma via VEGFA-mediated ERK pathway.Exp Cell Res. 2022 Sep 1;418(1):113267. doi: 10.1016/j.yexcr.2022.113267. Epub 2022 Jun 22. Exp Cell Res. 2022. PMID: 35752346 Review.
Cited by
-
Novel Insights into miR-944 in Cancer.Cancers (Basel). 2022 Aug 31;14(17):4232. doi: 10.3390/cancers14174232. Cancers (Basel). 2022. PMID: 36077769 Free PMC article. Review.
-
Cancer stem cells in brain tumors: From origin to clinical implications.MedComm (2020). 2023 Aug 9;4(4):e341. doi: 10.1002/mco2.341. eCollection 2023 Aug. MedComm (2020). 2023. PMID: 37576862 Free PMC article. Review.
-
MicroRNAs in Tumor Endothelial Cells: Regulation, Function and Therapeutic Applications.Cells. 2023 Jun 22;12(13):1692. doi: 10.3390/cells12131692. Cells. 2023. PMID: 37443725 Free PMC article.
-
Non-coding RNAs and glioma: Focus on cancer stem cells.Mol Ther Oncolytics. 2022 Sep 17;27:100-123. doi: 10.1016/j.omto.2022.09.005. eCollection 2022 Dec 15. Mol Ther Oncolytics. 2022. PMID: 36321132 Free PMC article. Review.
-
Tumor microenvironment of cancer stem cells: Perspectives on cancer stem cell targeting.Genes Dis. 2023 Jul 19;11(3):101043. doi: 10.1016/j.gendis.2023.05.024. eCollection 2024 May. Genes Dis. 2023. PMID: 38292177 Free PMC article. Review.
References
-
- Chang Y, Wu Q, Tian T, Li L, Guo X, Feng Z, Zhou J, Zhang L, Zhou S, Feng G, Han F, Yang J, Huang F. The influence of SRPK1 on glioma apoptosis, metastasis, and angiogenesis through the PI3K/Akt signaling pathway under normoxia. Tumour Biol. 2015; 36:6083–93. 10.1007/s13277-015-3289-2 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

