Down-regulation of A20 promotes immune escape of lung adenocarcinomas
- PMID: 34233950
- PMCID: PMC7611502
- DOI: 10.1126/scitranslmed.abc3911
Down-regulation of A20 promotes immune escape of lung adenocarcinomas
Abstract
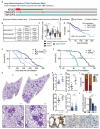
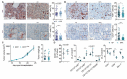
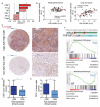
Inflammation is a well-known driver of lung tumorigenesis. One strategy by which tumor cells escape tight homeostatic control is by decreasing the expression of the potent anti-inflammatory protein tumor necrosis factor alpha-induced protein 3 (TNFAIP3), also known as A20. We observed that tumor cell intrinsic loss of A20 markedly enhanced lung tumorigenesis and was associated with reduced CD8+ T cell-mediated immune surveillance in patients with lung cancer and in mouse models. In mice, we observed that this effect was completely dependent on increased cellular sensitivity to interferon-γ (IFN-γ) signaling by aberrant activation of TANK-binding kinase 1 (TBK1) and increased downstream expression and activation of signal transducer and activator of transcription 1 (STAT1). Interrupting this autocrine feed forward loop by knocking out IFN-α/β receptor completely restored infiltration of cytotoxic T cells and rescued loss of A20 depending tumorigenesis. Downstream of STAT1, programmed death ligand 1 (PD-L1) was highly expressed in A20 knockout lung tumors. Accordingly, immune checkpoint blockade (ICB) treatment was highly efficient in mice harboring A20-deficient lung tumors. Furthermore, an A20 loss-of-function gene expression signature positively correlated with survival of melanoma patients treated with anti-programmed cell death protein 1. Together, we have identified A20 as a master immune checkpoint regulating the TBK1-STAT1-PD-L1 axis that may be exploited to improve ICB therapy in patients with lung adenocarcinoma.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
The authors declare no competing interests
Figures



References
-
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. - PubMed
-
- Prelaj A, Tay R, Ferrara R, Chaput N, Besse B, Califano R. Predictive biomarkers of response for immune checkpoint inhibitors in non-small-cell lung cancer. European journal of cancer (Oxford, England : 1990) 2019;106:144–159. - PubMed
-
- Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, Carcereny E, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. The New England journal of medicine. 2015;372:2018–2028. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

