Reply to: "Re-evaluating the evidence for a universal genetic boundary among microbial species"
- PMID: 34234115
- PMCID: PMC8263725
- DOI: 10.1038/s41467-021-24129-1
Reply to: "Re-evaluating the evidence for a universal genetic boundary among microbial species"
Conflict of interest statement
The authors declare no competing interests.
Figures
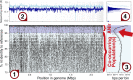
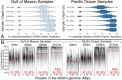
Comment on
-
High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries.Nat Commun. 2018 Nov 30;9(1):5114. doi: 10.1038/s41467-018-07641-9. Nat Commun. 2018. PMID: 30504855 Free PMC article.
-
Re-evaluating the evidence for a universal genetic boundary among microbial species.Nat Commun. 2021 Jul 7;12(1):4059. doi: 10.1038/s41467-021-24128-2. Nat Commun. 2021. PMID: 34234129 Free PMC article. No abstract available.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

