The epidemiology of Plasmodium vivax among adults in the Democratic Republic of the Congo
- PMID: 34234124
- PMCID: PMC8263614
- DOI: 10.1038/s41467-021-24216-3
The epidemiology of Plasmodium vivax among adults in the Democratic Republic of the Congo
Abstract
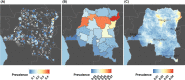
Reports of P. vivax infections among Duffy-negative hosts have accumulated throughout sub-Saharan Africa. Despite this growing body of evidence, no nationally representative epidemiological surveys of P. vivax in sub-Saharan Africa have been performed. To overcome this gap in knowledge, we screened over 17,000 adults in the Democratic Republic of the Congo (DRC) for P. vivax using samples from the 2013-2014 Demographic Health Survey. Overall, we found a 2.97% (95% CI: 2.28%, 3.65%) prevalence of P. vivax infections across the DRC. Infections were associated with few risk-factors and demonstrated a relatively flat distribution of prevalence across space with focal regions of relatively higher prevalence in the north and northeast. Mitochondrial genomes suggested that DRC P. vivax were distinct from circulating non-human ape strains and an ancestral European P. vivax strain, and instead may be part of a separate contemporary clade. Our findings suggest P. vivax is diffusely spread across the DRC at a low prevalence, which may be associated with long-term carriage of low parasitemia, frequent relapses, or a general pool of infections with limited forward propagation.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- WHO Team: Global Malaria Programme. World Malaria Report 2020. https://www.who.int/publications/i/item/9789240015791 (2020).
-
- Miller, L. H., Mason, S. J. & Clyde, D. F. The resistance factor to Plasmodium vivax in blacks: the Duffy-blood-group genotype, FyFy. N. Engl. J.295, 302–4(1976). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

