Theranostic cells: emerging clinical applications of synthetic biology
- PMID: 34234299
- PMCID: PMC8261392
- DOI: 10.1038/s41576-021-00383-3
Theranostic cells: emerging clinical applications of synthetic biology
Abstract
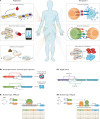
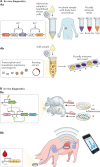
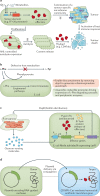
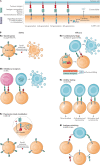
Synthetic biology seeks to redesign biological systems to perform novel functions in a predictable manner. Recent advances in bacterial and mammalian cell engineering include the development of cells that function in biological samples or within the body as minimally invasive diagnostics or theranostics for the real-time regulation of complex diseased states. Ex vivo and in vivo cell-based biosensors and therapeutics have been developed to target a wide range of diseases including cancer, microbiome dysbiosis and autoimmune and metabolic diseases. While probiotic therapies have advanced to clinical trials, chimeric antigen receptor (CAR) T cell therapies have received regulatory approval, exemplifying the clinical potential of cellular therapies. This Review discusses preclinical and clinical applications of bacterial and mammalian sensing and drug delivery platforms as well as the underlying biological designs that could enable new classes of cell diagnostics and therapeutics. Additionally, we describe challenges that must be overcome for more rapid and safer clinical use of engineered systems.
© 2021. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

