Scaling up reproducible research for single-cell transcriptomics using MetaNeighbor
- PMID: 34234317
- PMCID: PMC8826496
- DOI: 10.1038/s41596-021-00575-5
Scaling up reproducible research for single-cell transcriptomics using MetaNeighbor
Abstract
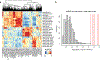
Single-cell RNA-sequencing data have significantly advanced the characterization of cell-type diversity and composition. However, cell-type definitions vary across data and analysis pipelines, raising concerns about cell-type validity and generalizability. With MetaNeighbor, we proposed an efficient and robust quantification of cell-type replicability that preserves dataset independence and is highly scalable compared to dataset integration. In this protocol, we show how MetaNeighbor can be used to characterize cell-type replicability by following a simple three-step procedure: gene filtering, neighbor voting and visualization. We show how these steps can be tailored to quantify cell-type replicability, determine gene sets that contribute to cell-type identity and pretrain a model on a reference taxonomy to rapidly assess newly generated data. The protocol is based on an open-source R package available from Bioconductor and GitHub, requires basic familiarity with Rstudio or the R command line and can typically be run in <5 min for millions of cells.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing financial interests
The authors declare that they have no competing financial interests.
Figures


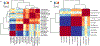












References
-
- Yao Z et al. An integrated transcriptomic and epigenomic atlas of mouse primary motor cortex cell types. bioRxiv 2020.02.29.970558 (2020) doi: 10.1101/2020.02.29.970558. - DOI
Related links
Key references using this protocol
Key data used in this protocol
-
- Yao Z et al. bioRxiv 2020.02.29.970558 (2020): 10.1101/2020.02.29.970558 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

