Impact of the Expression System on Recombinant Protein Production in Escherichia coli BL21
- PMID: 34234760
- PMCID: PMC8257044
- DOI: 10.3389/fmicb.2021.682001
Impact of the Expression System on Recombinant Protein Production in Escherichia coli BL21
Abstract
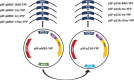
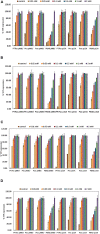
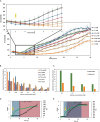
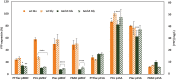
Recombinant protein production for medical, academic, or industrial applications is essential for our current life. Recombinant proteins are obtained mainly through microbial fermentation, with Escherichia coli being the host most used. In spite of that, some problems are associated with the production of recombinant proteins in E. coli, such as the formation of inclusion bodies, the metabolic burden, or the inefficient translocation/transport system of expressed proteins. Optimizing transcription of heterologous genes is essential to avoid these drawbacks and develop competitive biotechnological processes. Here, expression of YFP reporter protein is evaluated under the control of four promoters of different strength (P T7 lac , Ptrc, Ptac, and PBAD) and two different replication origins (high copy number pMB1' and low copy number p15A). In addition, the study has been carried out with the E. coli BL21 wt and the ackA mutant strain growing in a rich medium with glucose or glycerol as carbon sources. Results showed that metabolic burden associated with transcription and translation of foreign genes involves a decrease in recombinant protein expression. It is necessary to find a balance between plasmid copy number and promoter strength to maximize soluble recombinant protein expression. The results obtained represent an important advance on the most suitable expression system to improve both the quantity and quality of recombinant proteins in bioproduction engineering.
Keywords: Escherichia coli; expression system; microbial factory; origin of replication; promoter; recombinant protein.
Copyright © 2021 Lozano Terol, Gallego-Jara, Sola Martínez, Martínez Vivancos, Cánovas Díaz and de Diego Puente.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

