Gut Bacteria Associated With Monochamus saltuarius (Coleoptera: Cerambycidae) and Their Possible Roles in Host Plant Adaptations
- PMID: 34234761
- PMCID: PMC8256174
- DOI: 10.3389/fmicb.2021.687211
Gut Bacteria Associated With Monochamus saltuarius (Coleoptera: Cerambycidae) and Their Possible Roles in Host Plant Adaptations
Abstract
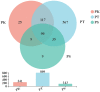
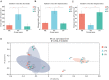
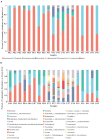
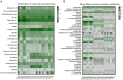
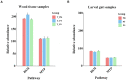
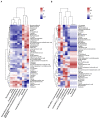
Monochamus saltuarius (Coleoptera: Cerambycidae) is an important native pest in the pine forests of northeast China and a dispersing vector of an invasive species Bursaphelenchus xylophilus. To investigate the bacterial gut diversity of M. saltuarius larvae in different host species, and infer the role of symbiotic bacteria in host adaptation, we used 16S rRNA gene Illumina sequencing and liquid chromatography-mass spectrometry metabolomics processing to obtain and compare the composition of the bacterial community and metabolites in the midguts of larvae feeding on three host tree species: Pinus koraiensis, Pinus sylvestris var. mongolica, and Pinus tabuliformis. Metabolomics in xylem samples from the three aforementioned hosts were also performed. Proteobacteria and Firmicutes were the predominant bacterial phyla in the larval gut. At the genus level, Klebsiella, unclassified_f__Enterobacteriaceae, Lactococcus, and Burkholderia-Caballeronia-Paraburkholderia were most dominant in P. koraiensis and P. sylvestris var. mongolica feeders, while Burkholderia-Caballeronia-Paraburkholderia, Dyella, Pseudoxanthomonas, and Mycobacterium were most dominant in P. tabuliformis feeders. Bacterial communities were similar in diversity in P. koraiensis and P. sylvestris var. mongolica feeders, while communities were highly diverse in P. tabuliformis feeders. Compared with the other two tree species, P. tabuliformis xylems had more diverse and abundant secondary metabolites, while larvae feeding on these trees had a stronger metabolic capacity for secondary metabolites than the other two host feeders. Correlation analysis of the association of microorganisms with metabolic features showed that dominant bacterial genera in P. tabuliformis feeders were more negatively correlated with plant secondary metabolites than those of other host tree feeders.
Keywords: borer; host adaptation; host-microbe interaction; intestinal bacterial composition; metabolomics; microbiota.
Copyright © 2021 Ge, Shi, Pei, Hou, Zong and Ren.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources

