Spore Germination of the Obligate Biotroph Spongospora subterranea: Transcriptome Analysis Reveals Germination Associated Genes
- PMID: 34234764
- PMCID: PMC8256667
- DOI: 10.3389/fmicb.2021.691877
Spore Germination of the Obligate Biotroph Spongospora subterranea: Transcriptome Analysis Reveals Germination Associated Genes
Abstract
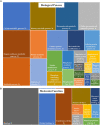
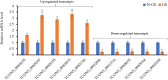
For soilborne pathogens, germination of the resting or dormant propagule that enables persistence within the soil environment is a key point in pathogenesis. Spongospora subterranea is an obligate soilborne protozoan that infects the roots and tubers of potato causing root and powdery scab disease for which there are currently no effective controls. A better understanding of the molecular basis of resting spore germination of S. subterranea could be important for development of novel disease interventions. However, as an obligate biotroph and soil dwelling organism, the application of new omics techniques for the study of the pre-infection process in S. subterranea has been problematic. Here, RNA sequencing was used to analyse the reprogramming of S. subterranea resting spores during the transition to zoospores in an in-vitro model. More than 63 million mean high-quality reads per sample were generated from the resting and germinating spores. By using a combination of reference-based and de novo transcriptome assembly, 6,664 unigenes were identified. The identified unigenes were subsequently annotated based on known proteins using BLAST search. Of 5,448 annotated genes, 570 genes were identified to be differentially expressed during the germination of S. subterranea resting spores, with most of the significant genes belonging to transcription and translation, amino acids biosynthesis, transport, energy metabolic processes, fatty acid metabolism, stress response and DNA repair. The datasets generated in this study provide a basic knowledge of the physiological processes associated with spore germination and will facilitate functional predictions of novel genes in S. subterranea and other plasmodiophorids. We introduce several candidate genes related to the germination of an obligate biotrophic soilborne pathogen which could be applied to the development of antimicrobial agents for soil inoculum management.
Keywords: Spongospora subterranea; obligate biotrophic; powdery scab; spore germination; transcriptomics.
Copyright © 2021 Balotf, Tegg, Nichols and Wilson.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Ah-Fong A. M., Kim K. S., Judelson H. S. (2017). RNA-seq of life stages of the oomycete Phytophthora infestans reveals dynamic changes in metabolic, signal transduction, and pathogenesis genes and a major role for calcium signaling in development. BMC Genom. 18:198. 10.1186/s12864-017-3585-x - DOI - PMC - PubMed
-
- Balendres M., Clark T., Tegg R., Wilson C. (2018). Germinate to exterminate: chemical stimulation of Spongospora subterranea resting spore germination and its potential to diminish soil inoculum. Plant Pathol. 67 902–908. 10.1111/ppa.12795 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

