Ultrafast and Reproducible Proteomics from Small Amounts of Heart Tissue Enabled by Azo and timsTOF Pro
- PMID: 34236868
- PMCID: PMC8349881
- DOI: 10.1021/acs.jproteome.1c00446
Ultrafast and Reproducible Proteomics from Small Amounts of Heart Tissue Enabled by Azo and timsTOF Pro
Abstract
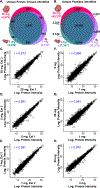
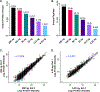
Global bottom-up mass spectrometry (MS)-based proteomics is widely used for protein identification and quantification to achieve a comprehensive understanding of the composition, structure, and function of the proteome. However, traditional sample preparation methods are time-consuming, typically including overnight tryptic digestion, extensive sample cleanup to remove MS-incompatible surfactants, and offline sample fractionation to reduce proteome complexity prior to online liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Thus, there is a need for a fast, robust, and reproducible method for protein identification and quantification from complex proteomes. Herein, we developed an ultrafast bottom-up proteomics method enabled by Azo, a photocleavable, MS-compatible surfactant that effectively solubilizes proteins and promotes rapid tryptic digestion, combined with the Bruker timsTOF Pro, which enables deeper proteome coverage through trapped ion mobility spectrometry (TIMS) and parallel accumulation-serial fragmentation (PASEF) of peptides. We applied this method to analyze the complex human cardiac proteome and identified nearly 4000 protein groups from as little as 1 mg of human heart tissue in a single one-dimensional LC-TIMS-MS/MS run with high reproducibility. Overall, we anticipate this ultrafast, robust, and reproducible bottom-up method empowered by both Azo and the timsTOF Pro will be generally applicable and greatly accelerate the throughput of large-scale quantitative proteomic studies. Raw data are available via the MassIVE repository with identifier MSV000087476.
Keywords: bottom-up proteomics; human heart proteomics; photocleavable surfactant; quantitative proteomics; sample preparation.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Gillet LC; Leitner A; Aebersold R Mass Spectrometry Applied to Bottom-Up Proteomics: Entering the High-Throughput Era for Hypothesis Testing. Annu. Rev. Anal. Chem. 2016, 9 (1), 449–472. - PubMed
-
- Aebersold R; Mann M Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537 (7620), 347–355. - PubMed
-
- Kuljanin M; Dieters-Castator DZ; Hess DA; Postovit L-M; Lajoie GA Comparison of sample preparation techniques for large-scale proteomics. Proteomics 2017, 17 (1–2), 1600337. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

