Listeria monocytogenes infection rewires host metabolism with regulatory input from type I interferons
- PMID: 34237114
- PMCID: PMC8266069
- DOI: 10.1371/journal.ppat.1009697
Listeria monocytogenes infection rewires host metabolism with regulatory input from type I interferons
Abstract
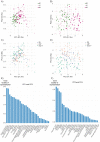
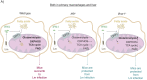
Listeria monocytogenes (L. monocytogenes) is a food-borne bacterial pathogen. Innate immunity to L. monocytogenes is profoundly affected by type I interferons (IFN-I). Here we investigated host metabolism in L. monocytogenes-infected mice and its potential control by IFN-I. Accordingly, we used animals lacking either the IFN-I receptor (IFNAR) or IRF9, a subunit of ISGF3, the master regulator of IFN-I-induced genes. Transcriptomes and metabolite profiles showed that L. monocytogenes infection induces metabolic rewiring of the liver. This affects various metabolic pathways including fatty acid (FA) metabolism and oxidative phosphorylation and is partially dependent on IFN-I signaling. Livers and macrophages from Ifnar1-/- mice employ increased glutaminolysis in an IRF9-independent manner, possibly to readjust TCA metabolite levels due to reduced FA oxidation. Moreover, FA oxidation inhibition provides protection from L. monocytogenes infection, explaining part of the protection of Irf9-/- and Ifnar1-/- mice. Our findings define a role of IFN-I in metabolic regulation during L. monocytogenes infection. Metabolic differences between Irf9-/- and Ifnar1-/- mice may underlie the different susceptibility of these mice against lethal infection with L. monocytogenes.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- WHO. WHO estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007–2015. WHO estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007–2015. 2015. p. 255. Available: https://www.who.int/foodsafety/publications/foodborne_disease/fergreport...
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

