SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19
- PMID: 34237248
- PMCID: PMC8139264
- DOI: 10.1016/j.chom.2021.05.010
SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19
Erratum in
-
SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19.Cell Host Microbe. 2022 Dec 14;30(12):1788. doi: 10.1016/j.chom.2022.10.017. Cell Host Microbe. 2022. PMID: 36521443 Free PMC article. No abstract available.
Abstract
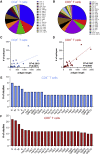
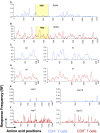
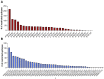
Over the past year, numerous studies in the peer reviewed and preprint literature have reported on the virological, epidemiological and clinical characteristics of the coronavirus, SARS-CoV-2. To date, 25 studies have investigated and identified SARS-CoV-2-derived T cell epitopes in humans. Here, we review these recent studies, how they were performed, and their findings. We review how epitopes identified throughout the SARS-CoV2 proteome reveal significant correlation between number of epitopes defined and size of the antigen provenance. We also report additional analysis of SARS-CoV-2 human CD4 and CD8 T cell epitope data compiled from these studies, identifying 1,400 different reported SARS-CoV-2 epitopes and revealing discrete immunodominant regions of the virus and epitopes that are more prevalently recognized. This remarkable breadth of epitope repertoire has implications for vaccine design, cross-reactivity, and immune escape by SARS-CoV-2 variants.
Keywords: CD4; CD8; SARS-CoV-2; T cells; epitopes.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.S. is a consultant for Gritstone, Flow Pharma, Merck, Epitogenesis, Gilead, and Avalia. S.C. is a consultant for Avalia. L.J.I. has filed for patent protection for various aspects of T cell epitope and vaccine design work.
Figures
References
-
- Abbas A.K., Lichtman A.H., Pillai S. Saunders Elsevier; 2007. Cellular and Molecular Immunology.
-
- Altmann D.M., Boyton R.J. SARS-CoV-2 T cell immunity: Specificity, function, durability, and role in protection. Sci. Immunol. 2020;5:5. - PubMed
-
- Assarsson E., Sidney J., Oseroff C., Pasquetto V., Bui H.H., Frahm N., Brander C., Peters B., Grey H., Sette A. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J. Immunol. 2007;178:7890–7901. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

