Predicting synchronized gene coexpression patterns from fibration symmetries in gene regulatory networks in bacteria
- PMID: 34238210
- PMCID: PMC8265036
- DOI: 10.1186/s12859-021-04213-5
Predicting synchronized gene coexpression patterns from fibration symmetries in gene regulatory networks in bacteria
Abstract
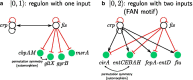
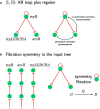
Background: Gene regulatory networks coordinate the expression of genes across physiological states and ensure a synchronized expression of genes in cellular subsystems, critical for the coherent functioning of cells. Here we address the question whether it is possible to predict gene synchronization from network structure alone. We have recently shown that synchronized gene expression can be predicted from symmetries in the gene regulatory networks described by the concept of symmetry fibrations. We showed that symmetry fibrations partition the genes into groups called fibers based on the symmetries of their 'input trees', the set of paths in the network through which signals can reach a gene. In idealized dynamic gene expression models, all genes in a fiber are perfectly synchronized, while less idealized models-with gene input functions differencing between genes-predict symmetry breaking and desynchronization.
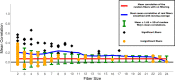
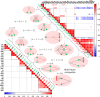
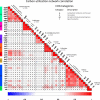
Results: To study the functional role of gene fibers and to test whether some of the fiber-induced coexpression remains in reality, we analyze gene fibrations for the gene regulatory networks of E. coli and B. subtilis and confront them with expression data. We find approximate gene coexpression patterns consistent with symmetry fibrations with idealized gene expression dynamics. This shows that network structure alone provides useful information about gene synchronization, and suggest that gene input functions within fibers may be further streamlined by evolutionary pressures to realize a coexpression of genes.
Conclusions: Thus, gene fibrations provide a sound conceptual tool to describe tunable coexpression induced by network topology and shaped by mechanistic details of gene expression.
Conflict of interest statement
The authors declare no competing interest.
Figures







Similar articles
-
Complexity reduction by symmetry: Uncovering the minimal regulatory network for logical computation in bacteria.PLoS Comput Biol. 2025 Apr 24;21(4):e1013005. doi: 10.1371/journal.pcbi.1013005. eCollection 2025 Apr. PLoS Comput Biol. 2025. PMID: 40273291 Free PMC article.
-
Fibration symmetries uncover the building blocks of biological networks.Proc Natl Acad Sci U S A. 2020 Apr 14;117(15):8306-8314. doi: 10.1073/pnas.1914628117. Epub 2020 Mar 31. Proc Natl Acad Sci U S A. 2020. PMID: 32234788 Free PMC article.
-
Complexity reduction by symmetry: uncovering the minimal regulatory network for logical computation in bacteria.ArXiv [Preprint]. 2025 Feb 25:arXiv:2310.10895v3. ArXiv. 2025. Update in: PLoS Comput Biol. 2025 Apr 24;21(4):e1013005. doi: 10.1371/journal.pcbi.1013005. PMID: 37904746 Free PMC article. Updated. Preprint.
-
Recent advances in gene function prediction using context-specific coexpression networks in plants.F1000Res. 2019 Feb 5;8:F1000 Faculty Rev-153. doi: 10.12688/f1000research.17207.1. eCollection 2019. F1000Res. 2019. PMID: 30800290 Free PMC article. Review.
-
Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders.Genes Brain Behav. 2014 Jan;13(1):13-24. doi: 10.1111/gbb.12106. Epub 2013 Dec 10. Genes Brain Behav. 2014. PMID: 24320616 Free PMC article. Review.
Cited by
-
Fibration symmetries and cluster synchronization in the Caenorhabditis elegans connectome.ArXiv [Preprint]. 2024 May 3:arXiv:2305.19367v2. ArXiv. 2024. Update in: PLoS One. 2024 Apr 10;19(4):e0297669. doi: 10.1371/journal.pone.0297669. PMID: 37396607 Free PMC article. Updated. Preprint.
-
Complexity reduction by symmetry: Uncovering the minimal regulatory network for logical computation in bacteria.PLoS Comput Biol. 2025 Apr 24;21(4):e1013005. doi: 10.1371/journal.pcbi.1013005. eCollection 2025 Apr. PLoS Comput Biol. 2025. PMID: 40273291 Free PMC article.
-
Fibration symmetry-breaking supports functional transitions in a brain network engaged in language.ArXiv [Preprint]. 2024 Sep 4:arXiv:2409.02674v1. ArXiv. 2024. PMID: 39279833 Free PMC article. Preprint.
-
Symmetries in metabolic networks of Escherichia coli.PNAS Nexus. 2025 Mar 17;4(3):pgaf080. doi: 10.1093/pnasnexus/pgaf080. eCollection 2025 Mar. PNAS Nexus. 2025. PMID: 40144777 Free PMC article.
-
Symmetries and synchronization from whole-neural activity in C. elegans connectome: Integration of functional and structural networks.ArXiv [Preprint]. 2024 Sep 4:arXiv:2409.02682v1. ArXiv. 2024. Update in: Proc Natl Acad Sci U S A. 2025 Jun 10;122(23):e2417850122. doi: 10.1073/pnas.2417850122. PMID: 39279832 Free PMC article. Updated. Preprint.
References
-
- Klipp E, Liebermeister W, Wierling C, Kowald A, Herwig R. Systems Biology: a textbook. Weinheim: Wiley-VCH; 2016.
-
- Palsson BØ. Systems biology: properties of reconstructed networks. New York: Cambridge University Press; 2006.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

