Venom-gland transcriptomic, venomic, and antivenomic profiles of the spine-bellied sea snake (Hydrophis curtus) from the South China Sea
- PMID: 34238212
- PMCID: PMC8268360
- DOI: 10.1186/s12864-021-07824-7
Venom-gland transcriptomic, venomic, and antivenomic profiles of the spine-bellied sea snake (Hydrophis curtus) from the South China Sea
Abstract
Background: A comprehensive evaluation of the -omic profiles of venom is important for understanding the potential function and evolution of snake venom. Here, we conducted an integrated multi-omics-analysis to unveil the venom-transcriptomic and venomic profiles in a same group of spine-bellied sea snakes (Hydrophis curtus) from the South China Sea, where the snake is a widespread species and might generate regionally-specific venom potentially harmful to human activities. The capacity of two heterologous antivenoms to immunocapture the H. curtus venom was determined for an in-depth evaluation of their rationality in treatment of H. curtus envenomation. In addition, a phylogenetic analysis by maximum likelihood was used to detect the adaptive molecular evolution of full-length toxin-coding unigenes.
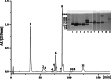
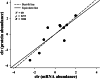
Results: A total of 90,909,384 pairs of clean reads were generated via Illumina sequencing from a pooled cDNA library of six specimens, and yielding 148,121 unigenes through de novo assembly. Sequence similarity searching harvested 63,845 valid annotations, including 63,789 non-toxin-coding and 56 toxin-coding unigenes belonging to 22 protein families. Three protein families, three-finger toxins (3-FTx), phospholipase A2 (PLA2), and cysteine-rich secretory protein, were detected in the venom proteome. 3-FTx (27.15% in the transcriptome/41.94% in the proteome) and PLA2 (59.71%/49.36%) were identified as the most abundant families in the venom-gland transcriptome and venom proteome. In addition, 24 unigenes from 11 protein families were shown to have experienced positive selection in their evolutionary history, whereas four were relatively conserved throughout evolution. Commercial Naja atra antivenom exhibited a stronger capacity than Bungarus multicinctus antivenom to immunocapture H. curtus venom components, especially short neurotoxins, with the capacity of both antivenoms to immunocapture short neurotoxins being weaker than that for PLA2s.
Conclusions: Our study clarified the venom-gland transcriptomic and venomic profiles along with the within-group divergence of a H. curtus population from the South China Sea. Adaptive evolution of most venom components driven by natural selection appeared to occur rapidly during evolutionary history. Notably, the utility of commercial N. atra and B. multicinctus antivenoms against H. curtus toxins was not comprehensive; thus, the development of species-specific antivenom is urgently needed.
Keywords: Antivenomic; Hydrophis curtus; Omics; Positive selection; Proteome; Snake venom; Transcriptome.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
The Venom of the Spine-Bellied Sea Snake (Hydrophis curtus): Proteome, Toxin Diversity and Intraspecific Variation.Int J Mol Sci. 2017 Dec 12;18(12):2695. doi: 10.3390/ijms18122695. Int J Mol Sci. 2017. PMID: 29231898 Free PMC article.
-
De Novo Venom-Gland Transcriptomics of Spine-Bellied Sea Snake (Hydrophis curtus) from Penang, Malaysia-Next-Generation Sequencing, Functional Annotation and Toxinological Correlation.Toxins (Basel). 2021 Feb 9;13(2):127. doi: 10.3390/toxins13020127. Toxins (Basel). 2021. PMID: 33572266 Free PMC article.
-
A comparative analysis of the proteomes and biological activities of the venoms from two sea snakes, Hydrophis curtus and Hydrophis cyanocinctus, from Hainan, China.Toxicon. 2020 Nov;187:35-46. doi: 10.1016/j.toxicon.2020.08.012. Epub 2020 Aug 29. Toxicon. 2020. PMID: 32871160
-
Biological and medical aspects related to the yellow-bellied sea snake Hydrophis platurus (Linnaeus, 1766): A view from Colombia.Travel Med Infect Dis. 2022 Sep-Oct;49:102410. doi: 10.1016/j.tmaid.2022.102410. Epub 2022 Aug 5. Travel Med Infect Dis. 2022. PMID: 35934312 Review.
-
Investigating Toxin Diversity and Abundance in Snake Venom Proteomes.Front Pharmacol. 2022 Jan 14;12:768015. doi: 10.3389/fphar.2021.768015. eCollection 2021. Front Pharmacol. 2022. PMID: 35095489 Free PMC article. Review.
Cited by
-
Bibliometric Analysis of Literature in Snake Venom-Related Research Worldwide (1933-2022).Animals (Basel). 2022 Aug 12;12(16):2058. doi: 10.3390/ani12162058. Animals (Basel). 2022. PMID: 36009648 Free PMC article.
-
Venom of the Annulated Sea Snake Hydrophis cyanocinctus: A Biochemically Simple but Genetically Complex Weapon.Toxins (Basel). 2021 Aug 6;13(8):548. doi: 10.3390/toxins13080548. Toxins (Basel). 2021. PMID: 34437419 Free PMC article.
-
DeTox: a pipeline for the detection of toxins in venomous organisms.Brief Bioinform. 2024 Jan 22;25(2):bbae094. doi: 10.1093/bib/bbae094. Brief Bioinform. 2024. PMID: 38493344 Free PMC article.
-
Marine Origin Ligands of Nicotinic Receptors: Low Molecular Compounds, Peptides and Proteins for Fundamental Research and Practical Applications.Biomolecules. 2022 Jan 23;12(2):189. doi: 10.3390/biom12020189. Biomolecules. 2022. PMID: 35204690 Free PMC article. Review.
-
Differences between Two Groups of Burmese Vipers (Viperidae: Azemiops) in the Proteomic Profiles, Immunoreactivity and Biochemical Functions of Their Venoms.Toxins (Basel). 2022 Aug 22;14(8):572. doi: 10.3390/toxins14080572. Toxins (Basel). 2022. PMID: 36006235 Free PMC article.
References
-
- Karthikeyan R, Balasubramamian T. Species diversity of sea snake (Hydrophiidae) distributed in the coramantal coast (East Coast of India) Int J Zool Res. 2007;3(3):107–131. doi: 10.3923/ijzr.2007.107.131. - DOI
-
- Calvete JJ, Ghezellou P, Paiva O, Matainaho T, Ghassempour A, Goudarzi H, Kraus F, Sanz L, Williams DJ. Snake venomics of two poorly known Hydrophiinae: comparative proteomics of the venoms of terrestrial Toxicocalamus longissimus and marine Hydrophis cyanocinctus. J Proteomics. 2012;75(13):4091–4101. doi: 10.1016/j.jprot.2012.05.026. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

