Computational analysis for identification of early diagnostic biomarkers and prognostic biomarkers of liver cancer based on GEO and TCGA databases and studies on pathways and biological functions affecting the survival time of liver cancer
- PMID: 34238253
- PMCID: PMC8268589
- DOI: 10.1186/s12885-021-08520-1
Computational analysis for identification of early diagnostic biomarkers and prognostic biomarkers of liver cancer based on GEO and TCGA databases and studies on pathways and biological functions affecting the survival time of liver cancer
Abstract
Background: Liver cancer is the sixth most commonly diagnosed cancer and the fourth most common cause of cancer death. The purpose of this work is to find new diagnostic biomarkers or prognostic biomarkers and explore the biological functions related to the prognosis of liver cancer.
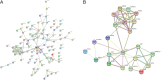
Methods: GSE25097 datasets were firstly obtained and compared with TCGA LICA datasets and an analysis of the overlapping differentially expressed genes (DEGs) was conducted. Cytoscape was used to screen out the Hub Genes among the DEGs. ROC curve analysis was used to screen the Hub Genes to determine the genes that could be used as diagnostic biomarkers. Kaplan-Meier analysis and Cox proportional hazards model screened genes associated with prognosis biomarkers, and further Gene Set Enrichment Analysis was performed on the prognosis genes to explore the mechanism affecting the survival and prognosis of liver cancer patients.
Results: 790 DEGs and 2162 DEGs were obtained respectively from the GSE25097 and TCGA LIHC data sets, and 102 Common DEGs were identified by overlapping the two DEGs. Further screening identified 22 Hub Genes from 102 Common DEGs. ROC and survival curves were used to analyze these 22 Hub Genes and it was found that there were 16 genes with a value of AUC > 90%. Among these, the expression levels of ESR1,SPP1 and FOSB genes were closely related to the survival time of liver cancer patients. Three common pathways of ESR1, FOBS and SPP1 genes were identified along with seven common pathways of ESR1 and SPP1 genes and four common pathways of ESR1 and FOSB genes.
Conclusions: SPP1, AURKA, NUSAP1, TOP2A, UBE2C, AFP, GMNN, PTTG1, RRM2, SPARCL1, CXCL12, FOS, DCN, SOCS3, FOSB and PCK1 can be used as diagnostic biomarkers for liver cancer, among which FOBS and SPP1 genes can also be used as prognostic biomarkers. Activation of the cell cycle-related pathway, pancreas beta cells pathway, and the estrogen signaling pathway, while on the other hand inhibition of the hallmark heme metabolism pathway, hallmark coagulation pathway, and the fat metabolism pathway may promote prognosis in liver cancer patients.
Keywords: Bioinformatics; Biomarker; Diagnostic biomarker; Liver cancer; Prognostic biomarker.
Conflict of interest statement
All authors declare that they have no conflict of interests.
Figures






References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68(6):394-424. - PubMed
-
- Piñero F, Tanno M, Aballay Soteras G, Tisi Baña M, Dirchwolf M, Fassio E, Ruf A, Mengarelli S, Borzi S, Fernández N, Ridruejo E, Descalzi V, Anders M, Mazzolini G, Reggiardo V, Marciano S, Perazzo F, Spina JC, McCormack L, Maraschio M, Lagues C, Gadano A, Villamil F, Silva M, Cairo F, Ameigeiras B, Argentinean Association for the Study of Liver Diseases (A.A.E.E.H). Argentinian clinical practice guideline for surveillance, diagnosis, staging and treatment of hepatocellular carcinoma. Ann Hepatol 2020;19(5):546–569. https://doi.org/10.1016/j.aohep.2020.06.003. - PubMed
-
- Feld JJ, Krassenburg LAP. What comes first: treatment of viral hepatitis or liver cancer? Dig Dis Sci 2019;64(4):1041–1049. https://doi.org/10.1007/s10620-019-05518-5. - PubMed
-
- Ringelhan M, O'Connor T, Protzer U, Heikenwalder M. The direct and indirect roles of HBV in liver cancer: prospective markers for HCC screening and potential therapeutic targets. J Pathol 2015;235(2):355–367. https://doi.org/10.1002/path.4434. - PubMed
-
- Pham C, Sin MK. Use of electronic health Records at Federally Qualified Health Centers: a potent tool to increase viral hepatitis screening and address the climbing incidence of liver Cancer. J Cancer Educ 2020. https://doi.org/10.1007/s13187-020-01741-1. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

