APOE genotype dependent molecular abnormalities in the cerebrovasculature of Alzheimer's disease and age-matched non-demented brains
- PMID: 34238312
- PMCID: PMC8268468
- DOI: 10.1186/s13041-021-00803-9
APOE genotype dependent molecular abnormalities in the cerebrovasculature of Alzheimer's disease and age-matched non-demented brains
Abstract
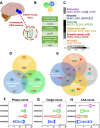
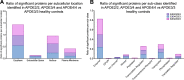
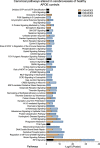
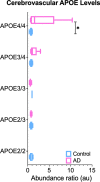
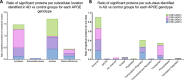
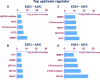
Cerebrovascular dysfunction is a hallmark feature of Alzheimer's disease (AD). One of the greatest risk factors for AD is the apolipoprotein E4 (E4) allele. The APOE4 genotype has been shown to negatively impact vascular amyloid clearance, however, its direct influence on the molecular integrity of the cerebrovasculature compared to other APOE variants (APOE2 and APOE3) has been largely unexplored. To address this, we employed a 10-plex tandem isobaric mass tag approach in combination with an ultra-high pressure liquid chromatography MS/MS (Q-Exactive) method, to interrogate unbiased proteomic changes in cerebrovessels from AD and healthy control brains with different APOE genotypes. We first interrogated changes between healthy control cases to identify underlying genotype specific effects in cerebrovessels. EIF2 signaling, regulation of eIF4 and 70S6K signaling and mTOR signaling were the top significantly altered pathways in E4/E4 compared to E3/E3 cases. Oxidative phosphorylation, EIF2 signaling and mitochondrial dysfunction were the top significant pathways in E2E2 vs E3/E3cases. We also identified AD-dependent changes and their interactions with APOE genotype and found the highest number of significant proteins from this interaction was observed in the E3/E4 (192) and E4/E4 (189) cases. As above, EIF2, mTOR signaling and eIF4 and 70S6K signaling were the top three significantly altered pathways in E4 allele carriers (i.e. E3/E4 and E4/E4 genotypes). Of all the cerebrovascular cell-type specific markers identified in our proteomic analyses, endothelial cell, astrocyte, and smooth muscle cell specific protein markers were significantly altered in E3/E4 cases, while endothelial cells and astrocyte specific protein markers were altered in E4/E4 cases. These proteomic changes provide novel insights into the longstanding link between APOE4 and cerebrovascular dysfunction, implicating a role for impaired autophagy, ER stress, and mitochondrial bioenergetics. These APOE4 dependent changes we identified could provide novel cerebrovascular targets for developing disease modifying strategies to mitigate the effects of APOE4 genotype on AD pathogenesis.
Keywords: APOE; Aging; Alzheimer’s disease; Cerebrovasculature; Endothelial cells; Mass spectrometry; Mural cells; Proteomics.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- de la Torre J. The vascular hypothesis of Alzheimer’s Disease: a key to preclinical prediction of dementia using neuroimaging. Mecocci P, editor. J Alzheimers Dis. 2018;63:35–52. 10.3233/JAD-180004 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

