Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade
- PMID: 34239137
- PMCID: PMC11107795
- DOI: 10.1038/s41591-021-01406-6
Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade
Abstract
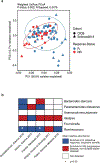
Treatment with combined immune checkpoint blockade (CICB) targeting CTLA-4 and PD-1 is associated with clinical benefit across tumor types, but also a high rate of immune-related adverse events. Insights into biomarkers and mechanisms of response and toxicity to CICB are needed. To address this, we profiled the blood, tumor and gut microbiome of 77 patients with advanced melanoma treated with CICB, with a high rate of any ≥grade 3 immune-related adverse events (49%) with parallel studies in pre-clinical models. Tumor-associated immune and genomic biomarkers of response to CICB were similar to those identified for ICB monotherapy, and toxicity from CICB was associated with a more diverse peripheral T-cell repertoire. Profiling of gut microbiota demonstrated a significantly higher abundance of Bacteroides intestinalis in patients with toxicity, with upregulation of mucosal IL-1β in patient samples of colitis and in pre-clinical models. Together, these data offer potential new therapeutic angles for targeting toxicity to CICB.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
M.C.A. reports advisory board participation and honoraria from Merck Sharp and Dohme, outside the submitted work. V.G. and Z.A.C. are currently employees of AstraZeneca. R.N.A. reports research funding from Bristol-Myers Squibb (BMS), Merck and Genentech, all outside the submitted work. R.C.P. reports honoraria EMD Serono, Merck, Norvartis and Pfizer as well as consulting for Merck, BMS, Novartis, Pfizer and AstraZeneca, and research funding from Merck and Novartis outside the submitted work. H.A.T. reports personal fees from Novartis, grants from Merck and Celgene and grants and personal fees from BMS and Genentech, all outside the submitted work. M.A.D. has been a consultant to Roche/Genentech, Array, Novartis, BMS, GlaxoSmithKline (GSK), Sanofi-Aventis, Vaccinex and Apexigen, and he has been the PI of research grants to UT MD Anderson by Roche/Genentech, GSK, Sanofi-Aventis, Merck, Myriad and Oncothyreon. W.-J.H. reports research grants from Merck, BMS, MedImmune and GSK and has served on an advisory board for Merck, all outside the submitted work. J.E.G. reports advisory board participation with Merck, Regeneron, BMS, Novartis and Syndax. A.J.L. reports personal fees from BMS, Novartis, Genentech/Roche and Merck; personal fees and non-financial support from ArcherDX and Beta-Cat; grants and non-financial support from Medimmune/AstraZeneca and Sanofi; grants, personal fees and non-financial support from Janssen, all outside the submitted work. M.T.T. reports personal fees from Myriad Genetics, Seattle Genetics and Novartis, all outside the submitted work. A.P.C. reports advisory roles and/or stock ownership for Immunai and Vastbiome. M.A.P. reports honoraria from BMS and Merck, consulting fees from BMS, Merck, Array BioPharma, Novartis, Incyte, NewLink Genetics, Aduro, Eisai and Pfizer and institutional support from RGenix, Infinity, BMS, Merck, Array BioPharma, Novartis and AstraZeneca. S.P.P. reports institutional support for a clinical trial from InxMed. J.L.M. reports honoraria from Roche, BMS and Merck. R.R.J. has consulted for Karius, Merck, Microbiome DX and Prolacta, and is on the scientific advisory boards of Kaleido, LISCure, Maat Pharma and Seres, and has received patent royalties licensed to Seres. P.S. reports consulting, advisory roles and/or stocks/ownership for Achelois, Adaptive Biotechnologies, Apricity Health, BioAlta, BioNTech, Codiak Biosciences, Constellation, Dragonfly Therapeutics, Forty-Seven Inc., Hummingbird, ImaginAb, Infinity Pharma, Jounce Therapeutics, Lave Therapeutics, Lytix Biopharma, Marker Therapeutics, Oncolytics, Phenomics and Polaris, and owns a patent licensed to Jounce Therapeutics. J.P.A. reports consulting, advisory roles and/or stocks/ownership for Achelois, Adaptive Biotechnologies, Apricity Health, BioAlta, BioNTech, Codiak Biosciences, Constellation, Dragonfly Therapeutics, Forty-Seven Inc., Hummingbird, ImaginAb, Jounce Therapeutics, Lave Therapeutics, Lytix Biopharma, Marker Therapeutics, Phenomics and Polaris, and owns a patent licensed to Jounce Therapeutics. B.R. reports advisory board membership for Vedanta and research funding from Vedanta, Davoltera and Kaleido. V.G., C.N.S., A.R. and J.A.W. are co-inventors on US patent PCT/US17/53,717, relating to the microbiome. J.A.W., V.G., M.C.A., L. Zitvogel and V.I. are co-inventors on a provisional US patent (WO2020106983A1) relating to the microbiome, relevant to the current work. L. Zitvogel is the main founder of EverImmune, a biotech company devoted to the use of commensal bacteria for the treatment of cancers, is on the board of administrators of Transgene and in the scientific advisory board of EpiVax, Lytix Biopharma, and has received research contracts from Kaleido, BMS, Incyte, Transgene, MERUS and GSK. J.A.W. reports speaker fees from Imedex, Dava Oncology, Omniprex, Illumina, Gilead, MedImmune and BMS; consultant/advisor roles or advisory board membership for Roche-Genentech, Novartis, AstraZeneca, GSK, BMS, Merck/MSD, Biothera Pharma and Microbiome DX; and receives clinical trial support from GSK, Roche-Genentech, BMS and Novartis, all outside the current work. The remaining authors declare no competing interests.
Figures










Comment in
-
Can gut microbes predict efficacy and toxicity of combined immune checkpoint blockade?Cancer Cell. 2021 Oct 11;39(10):1314-1316. doi: 10.1016/j.ccell.2021.09.013. Cancer Cell. 2021. PMID: 34637746
References
-
- Sznol M et al. Pooled analysis safety profile of nivolumab and ipilimumab combination therapy in patients with advanced melanoma. J. Clin. Oncol 35, 3815–3822 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

