Mechanisms of Genome Maintenance in Plants: Playing It Safe With Breaks and Bumps
- PMID: 34239541
- PMCID: PMC8258418
- DOI: 10.3389/fgene.2021.675686
Mechanisms of Genome Maintenance in Plants: Playing It Safe With Breaks and Bumps
Abstract
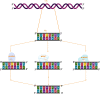
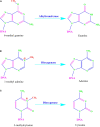
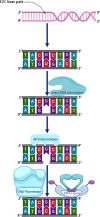
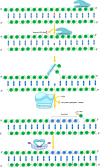
Maintenance of genomic integrity is critical for the perpetuation of all forms of life including humans. Living organisms are constantly exposed to stress from internal metabolic processes and external environmental sources causing damage to the DNA, thereby promoting genomic instability. To counter the deleterious effects of genomic instability, organisms have evolved general and specific DNA damage repair (DDR) pathways that act either independently or mutually to repair the DNA damage. The mechanisms by which various DNA repair pathways are activated have been fairly investigated in model organisms including bacteria, fungi, and mammals; however, very little is known regarding how plants sense and repair DNA damage. Plants being sessile are innately exposed to a wide range of DNA-damaging agents both from biotic and abiotic sources such as ultraviolet rays or metabolic by-products. To escape their harmful effects, plants also harbor highly conserved DDR pathways that share several components with the DDR machinery of other organisms. Maintenance of genomic integrity is key for plant survival due to lack of reserve germline as the derivation of the new plant occurs from the meristem. Untowardly, the accumulation of mutations in the meristem will result in a wide range of genetic abnormalities in new plants affecting plant growth development and crop yield. In this review, we will discuss various DNA repair pathways in plants and describe how the deficiency of each repair pathway affects plant growth and development.
Keywords: DNA damage; DNA repair pathways; DNA replication; genome integrity; mutations.
Copyright © 2021 Raina, Sahu, Laskar, Rajora, Sao, Khan and Ganai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Ahmad A., Nay S. L., O’Connor T. R. (2015). “Direct reversal repair in mammalian cells,” Advances in DNA Repair ed. Chen C. (IntechOpen; ), 95–128.
Publication types
LinkOut - more resources
Full Text Sources

