The Research on the Treatment of Metastatic Skin Cutaneous Melanoma by Huanglian Jiedu Decoction Based on the Analysis of Immune Infiltration Analysis
- PMID: 34239596
- PMCID: PMC8241506
- DOI: 10.1155/2021/9952060
The Research on the Treatment of Metastatic Skin Cutaneous Melanoma by Huanglian Jiedu Decoction Based on the Analysis of Immune Infiltration Analysis
Abstract
Objective: To explore the potential mechanism of Huanglian Jiedu Decoction (HJD) treatment and prevention of metastatic Cutaneous Melanoma (CM) occurrence and metastasis based on network pharmacological methods and immune infiltration analysis.
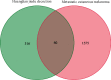
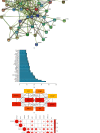
Methods: The GEO database was used to obtain metastatic CM disease targets, the TCMSP database and the HERB database were used to obtain HJD action targets, core genes were screened by protein interaction network, and the potential mechanism of HJD in the treatment of metastatic CM was explored by enrichment analysis, prognostic analysis and immune infiltration analysis.
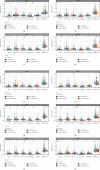
Results: HJD treatment of metastatic CM involved 60 targets, enrichment analysis showed that HJD treatment of metastatic CM involved Chemokine signaling pathway, NF-kappa B signaling pathway, and Fluid shear stress and atherosclerosis, etc. Prognostic analysis revealed that HJD had a certain ability to improve the prognosis of metastatic CM patients. Immune infiltration analysis showed that HJD could inhibit the immune cell infiltration of metastatic CM patients by acting on related targets.
Conclusions: Our study identified the potential mechanism of HJD in the treatment of metastatic CM through network pharmacology, and revealed the mechanism of HJD in the prevention of Skin Cutaneous Melanoma metastasis through immune infiltration analysis and prognostic analysis.
Copyright © 2021 Ding Li et al.
Conflict of interest statement
The authors declare that they have no conflicts of interests.
Figures






References
-
- Gómez-Abenza E., Ibáñez-Molero S., García-Moreno D., et al. Zebrafish modeling reveals that SPINT1 regulates the aggressiveness of skin cutaneous melanoma and its crosstalk with tumor immune microenvironment. Journal of Experimental & Clinical Cancer Research. 2019;38(1):p. 405. doi: 10.1186/s13046-019-1389-3. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

