Pan-Cancer Analysis of the Prognostic and Immunological Role of HSF1: A Potential Target for Survival and Immunotherapy
- PMID: 34239690
- PMCID: PMC8238600
- DOI: 10.1155/2021/5551036
Pan-Cancer Analysis of the Prognostic and Immunological Role of HSF1: A Potential Target for Survival and Immunotherapy
Abstract
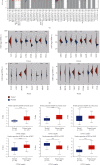
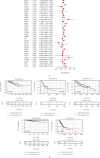
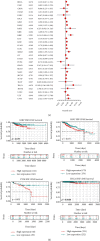
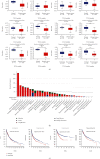
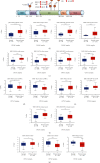
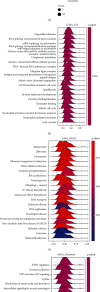
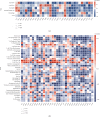
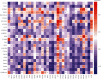
Emerging evidence revealed the significant roles of heat shock factor 1 (HSF1) in cancer initiation, development, and progression, but there is no pan-cancer analysis of HSF1. The present study first comprehensively investigated the expression profiles and prognostic significance of HSF1 and the relationship of HSF1 with clinicopathological parameters and immune cell infiltration using bioinformatic techniques. HSF1 is significantly upregulated in various common cancers, and it is associated with prognosis. Pan-cancer Cox regression analysis indicated that the high expression of HSF1 was associated with poor overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI) in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), head and neck squamous cell carcinoma (HNSC), and kidney renal papillary cell carcinoma (KIRP) patients. The methylation of HSF1 DNA was decreased in most cancers and negatively correlated with the HSF1 expression. Increased phosphorylation of S303, S307, and S363 in HSF1 was observed in some cancers. HSF1 remarkably correlated with the levels of infiltrating cells and immune checkpoint genes. Our pan-cancer analysis provides a deep understanding of the functions of HSF1 in oncogenesis and metastasis in different cancers.
Copyright © 2021 Fei Chen et al.
Conflict of interest statement
The authors declare no conflicts.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

