Understanding small ORF diversity through a comprehensive transcription feature classification
- PMID: 34240112
- PMCID: PMC8435553
- DOI: 10.1093/dnares/dsab007
Understanding small ORF diversity through a comprehensive transcription feature classification
Abstract
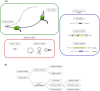
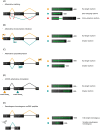
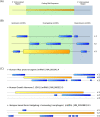
Small open reading frames (small ORFs/sORFs/smORFs) are potentially coding sequences smaller than 100 codons that have historically been considered junk DNA by gene prediction software and in annotation screening; however, the advent of next-generation sequencing has contributed to the deeper investigation of junk DNA regions and their transcription products, resulting in the emergence of smORFs as a new focus of interest in systems biology. Several smORF peptides were recently reported in non-canonical mRNAs as new players in numerous biological contexts; however, their relevance is still overlooked in coding potential analysis. Hence, this review proposes a smORF classification based on transcriptional features, discussing the most promising approaches to investigate smORFs based on their different characteristics. First, smORFs were divided into non-expressed (intergenic) and expressed (genic) smORFs. Second, genic smORFs were classified as smORFs located in non-coding RNAs (ncRNAs) or canonical mRNAs. Finally, smORFs in ncRNAs were further subdivided into sequences located in small or long RNAs, whereas smORFs located in canonical mRNAs were subdivided into several specific classes depending on their localization along the gene. We hope that this review provides new insights into large-scale annotations and reinforces the role of smORFs as essential components of a hidden coding DNA world.
Keywords: alternative ORFs; dual functional RNA; genome annotation; long non-coding RNA; smORF peptides.
© The Author(s) 2021. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

