Global Genetic Heterogeneity in Adaptive Traits
- PMID: 34240182
- PMCID: PMC8557469
- DOI: 10.1093/molbev/msab208
Global Genetic Heterogeneity in Adaptive Traits
Abstract
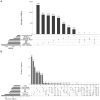
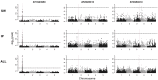
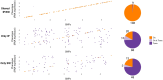
Understanding the genetic architecture of complex traits is a major objective in biology. The standard approach for doing so is genome-wide association studies (GWAS), which aim to identify genetic polymorphisms responsible for variation in traits of interest. In human genetics, consistency across studies is commonly used as an indicator of reliability. However, if traits are involved in adaptation to the local environment, we do not necessarily expect reproducibility. On the contrary, results may depend on where you sample, and sampling across a wide range of environments may decrease the power of GWAS because of increased genetic heterogeneity. In this study, we examine how sampling affects GWAS in the model plant species Arabidopsis thaliana. We show that traits like flowering time are indeed influenced by distinct genetic effects in local populations. Furthermore, using gene expression as a molecular phenotype, we show that some genes are globally affected by shared variants, whereas others are affected by variants specific to subpopulations. Remarkably, the former are essentially all cis-regulated, whereas the latter are predominately affected by trans-acting variants. Our result illustrate that conclusions about genetic architecture can be extremely sensitive to sampling and population structure.
Keywords: GWAS; evolutionary genomics; genetic architecture; regulation of gene expression.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Ågren J, Oakley CG, Lundemo S, Schemske DW.. 2017. Adaptive divergence in flowering time among natural populations of Arabidopsis thaliana: estimates of selection and QTL mapping. Evolution 71(3):550–564. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

