How subtle changes in 3D structure can create large changes in transcription
- PMID: 34240703
- PMCID: PMC8352591
- DOI: 10.7554/eLife.64320
How subtle changes in 3D structure can create large changes in transcription
Abstract
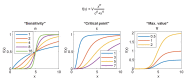
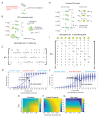
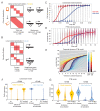
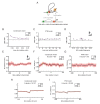
Animal genomes are organized into topologically associated domains (TADs). TADs are thought to contribute to gene regulation by facilitating enhancer-promoter (E-P) contacts within a TAD and preventing these contacts across TAD borders. However, the absolute difference in contact frequency across TAD boundaries is usually less than 2-fold, even though disruptions of TAD borders can change gene expression by 10-fold. Existing models fail to explain this hypersensitive response. Here, we propose a futile cycle model of enhancer-mediated regulation that can exhibit hypersensitivity through bistability and hysteresis. Consistent with recent experiments, this regulation does not exhibit strong correlation between E-P contact and promoter activity, even though regulation occurs through contact. Through mathematical analysis and stochastic simulation, we show that this system can create an illusion of E-P biochemical specificity and explain the importance of weak TAD boundaries. It also offers a mechanism to reconcile apparently contradictory results from recent global TAD disruption with local TAD boundary deletion experiments. Together, these analyses advance our understanding of cis-regulatory contacts in controlling gene expression and suggest new experimental directions.
Keywords: 3D genome; D. melanogaster; TAD; chromosomes; computational biology; gene expression; human; mouse; stochastic modeling; systems biology; transcription.
© 2021, Xiao et al.
Conflict of interest statement
JX, AH, AB No competing interests declared
Figures






References
-
- Alberts B, Johnson A, Lewis J, Morgan D, Raff M, Keith Roberts PW O. Molecular Biology of the Cell. W. W. Norton & Company; 2018.
-
- Allahyar A, Vermeulen C, Bouwman BAM, Krijger PHL, Verstegen M, Geeven G, van Kranenburg M, Pieterse M, Straver R, Haarhuis JHI, Jalink K, Teunissen H, Renkens IJ, Kloosterman WP, Rowland BD, de Wit E, de Ridder J, de Laat W. Enhancer hubs and loop collisions identified from single-allele topologies. Nature Genetics. 2018;50:1151–1160. doi: 10.1038/s41588-018-0161-5. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

