Antibody landscape against SARS-CoV-2 reveals significant differences between non-structural/accessory and structural proteins
- PMID: 34242574
- PMCID: PMC8233850
- DOI: 10.1016/j.celrep.2021.109391
Antibody landscape against SARS-CoV-2 reveals significant differences between non-structural/accessory and structural proteins
Abstract
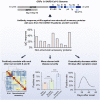
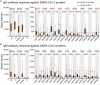
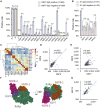
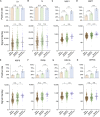
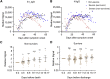
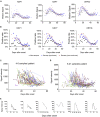
The immunogenicity of the SARS-CoV-2 proteome is largely unknown, especially for non-structural proteins and accessory proteins. In this study, we collect 2,360 COVID-19 sera and 601 control sera. We analyze these sera on a protein microarray with 20 proteins of SARS-CoV-2, building an antibody response landscape for immunoglobulin (Ig)G and IgM. Non-structural proteins and accessory proteins NSP1, NSP7, NSP8, RdRp, ORF3b, and ORF9b elicit prevalent IgG responses. The IgG patterns and dynamics of non-structural/accessory proteins are different from those of the S and N proteins. The IgG responses against these six proteins are associated with disease severity and clinical outcome, and they decline sharply about 20 days after symptom onset. In non-survivors, a sharp decrease of IgG antibodies against S1 and N proteins before death is observed. The global antibody responses to non-structural/accessory proteins revealed here may facilitate a deeper understanding of SARS-CoV-2 immunology.
Keywords: COVID-19; SARS-CoV-2; humoral immunity; immune response; non-structural/accessory proteins; proteome microarray.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
References
-
- Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O. The coding capacity of SARS-CoV-2. Nature. 2021;589:125–130. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

