Generation of caudal-type serotonin neurons and hindbrain-fate organoids from hPSCs
- PMID: 34242615
- PMCID: PMC8365029
- DOI: 10.1016/j.stemcr.2021.06.006
Generation of caudal-type serotonin neurons and hindbrain-fate organoids from hPSCs
Abstract
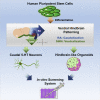
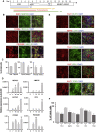
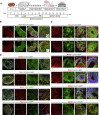
Serotonin (5-HT) neurons, the major components of the raphe nuclei, arise from ventral hindbrain progenitors. Based on anatomical location and axonal projection, 5-HT neurons are coarsely divided into rostral and caudal groups. Here, we propose a novel strategy to generate hindbrain 5-HT neurons from human pluripotent stem cells (hPSCs), which involves the formation of ventral-type neural progenitor cells and stimulation of the hindbrain 5-HT neural development. A caudalizing agent, retinoid acid, was used to direct the cells into the hindbrain cell fate. Approximately 30%-40% of hPSCs successfully developed into 5-HT-expressing neurons using our protocol, with the majority acquiring a caudal rhombomere identity (r5-8). We further modified our monolayer differentiation system to generate 5-HT neuron-enriched hindbrain-like organoids. We also suggest downstream applications of our 5-HT monolayer and organoid cultures to study neuronal response to gut microbiota. Our methodology could become a powerful tool for future studies related to 5-HT neurotransmission.
Keywords: hindbrain; human pluripotent stem cells; neuronal development; organoids; serotonin neurons.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Allen Institute for Brain Science . internet resource; 2010. Allen Human Brain Atlas.
-
- Briscoe J., Ericson J. The specification of neuronal identity by graded sonic hedgehog signalling. Semin. Cell Dev. Biol. 1999;10:353–362. - PubMed
-
- Brodski C., Weisenhorn D.M.V., Signore M., Sillaber I., Oesterheld M., Broccoli V., Acampora D., Simeone A., Wurst W. Location and size of dopaminergic and serotonergic cell populations are controlled by the position of the midbrain-hindbrain organizer. J. Neurosci. 2003;23:4199–4207. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

