Genome wide association study of agronomic and seed traits in a world collection of proso millet (Panicum miliaceum L.)
- PMID: 34243721
- PMCID: PMC8268170
- DOI: 10.1186/s12870-021-03111-5
Genome wide association study of agronomic and seed traits in a world collection of proso millet (Panicum miliaceum L.)
Abstract
Background: The climate crisis threatens sustainability of crop production worldwide. Crop diversification may enhance food security while reducing the negative impacts of climate change. Proso millet (Panicum milaceum L.) is a minor cereal crop which holds potential for diversification and adaptation to different environmental conditions. In this study, we assembled a world collection of proso millet consisting of 88 varieties and landraces to investigate its genomic and phenotypic diversity for seed traits, and to identify marker-trait associations (MTA).
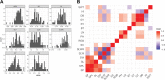
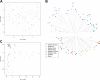
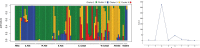
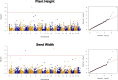
Results: Sequencing of restriction-site associated DNA fragments yielded 494 million reads and 2,412 high quality single nucleotide polymorphisms (SNPs). SNPs were used to study the diversity in the collection and perform a genome wide association study (GWAS). A genotypic diversity analysis separated accessions originating in Western Europe, Eastern Asia and Americas from accessions sampled in Southern Asia, Western Asia, and Africa. A Bayesian structure analysis reported four cryptic genetic groups, showing that landraces accessions had a significant level of admixture and that most of the improved proso millet materials clustered separately from landraces. The collection was highly diverse for seed traits, with color varying from white to dark brown and width spanning from 1.8 to 2.6 mm. A GWAS study for seed morphology traits identified 10 MTAs. In addition, we identified three MTAs for agronomic traits that were previously measured on the collection.
Conclusion: Using genomics and automated seed phenotyping, we elucidated phylogenetic relationships and seed diversity in a global millet collection. Overall, we identified 13 MTAs for key agronomic and seed traits indicating the presence of alleles with potential for application in proso breeding programs.
Keywords: Agrobiodiversity; GWAS; NUC; Population genetics; Proso millet; Seed morphology.
© 2021. The Author(s).
Conflict of interest statement
Authors declare no competing interest.
Figures
Similar articles
-
Genome-wide assessment of population structure and association mapping for agronomic and grain nutritional traits in proso millet (Panicum miliaceum L.).Sci Rep. 2024 Sep 19;14(1):21920. doi: 10.1038/s41598-024-72319-w. Sci Rep. 2024. PMID: 39300236 Free PMC article.
-
Genome-wide association study reveals marker-trait associations for major agronomic traits in proso millet (Panicum miliaceum L.).Planta. 2024 Jul 4;260(2):44. doi: 10.1007/s00425-024-04465-4. Planta. 2024. PMID: 38963439
-
Evaluation of the agronomic traits and correlation analysis of phenotypes of proso millet (Panicum miliaceum L.) germplasm in Kazakhstan.Braz J Biol. 2024 Nov 25;84:e287947. doi: 10.1590/1519-6984.287947. eCollection 2024. Braz J Biol. 2024. PMID: 39607246
-
Proso Millet (Panicum miliaceum L.) and Its Potential for Cultivation in the Pacific Northwest, U.S.: A Review.Front Plant Sci. 2017 Jan 9;7:1961. doi: 10.3389/fpls.2016.01961. eCollection 2016. Front Plant Sci. 2017. PMID: 28119699 Free PMC article. Review.
-
Genetic diversity and genomic resources available for the small millet crops to accelerate a New Green Revolution.Front Plant Sci. 2015 Mar 24;6:157. doi: 10.3389/fpls.2015.00157. eCollection 2015. Front Plant Sci. 2015. PMID: 25852710 Free PMC article. Review.
Cited by
-
Mineral, seed morphology, and agronomic characteristics of proso millet grown in the inland Pacific Northwest.Front Nutr. 2024 Sep 11;11:1394136. doi: 10.3389/fnut.2024.1394136. eCollection 2024. Front Nutr. 2024. PMID: 39323567 Free PMC article.
-
SNP discovery in proso millet (Panicum miliaceum L.) using low-pass genome sequencing.Plant Direct. 2022 Sep 13;6(9):e447. doi: 10.1002/pld3.447. eCollection 2022 Sep. Plant Direct. 2022. PMID: 36176305 Free PMC article.
-
Genomic resources, opportunities, and prospects for accelerated improvement of millets.Theor Appl Genet. 2024 Nov 20;137(12):273. doi: 10.1007/s00122-024-04777-9. Theor Appl Genet. 2024. PMID: 39565376 Free PMC article. Review.
-
A Review on Isolation, Characterization, Modification, and Applications of Proso Millet Starch.Foods. 2023 Jun 19;12(12):2413. doi: 10.3390/foods12122413. Foods. 2023. PMID: 37372623 Free PMC article. Review.
-
Millets: The future crops for the tropics - Status, challenges and future prospects.Heliyon. 2023 Nov 10;9(11):e22123. doi: 10.1016/j.heliyon.2023.e22123. eCollection 2023 Nov. Heliyon. 2023. PMID: 38058626 Free PMC article. Review.
References
-
- Das IK, Rakshit S. Chapter 1 - Millets, their importance, and production constraints. In: Das IK, Padmaja PG, editors. Biotic stress resistance in millets. Amsterdam: Academic Press, Elsevier; 2016. p. 3–19.
-
- Lágler R, Gyulai G, Humphreys M, Szabó Z, Horváth L, Bittsánszky A, et al. Morphological and molecular analysis of common millet (P. miliaceum) cultivars compared to an aDNA sample from the 15th century (Hungary) Euphytica. 2005;146(1):77–85. doi: 10.1007/s10681-005-5814-7. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

