Genetic prediction of complex traits with polygenic scores: a statistical review
- PMID: 34243982
- PMCID: PMC8511058
- DOI: 10.1016/j.tig.2021.06.004
Genetic prediction of complex traits with polygenic scores: a statistical review
Abstract
Accurate genetic prediction of complex traits can facilitate disease screening, improve early intervention, and aid in the development of personalized medicine. Genetic prediction of complex traits requires the development of statistical methods that can properly model polygenic architecture and construct a polygenic score (PGS). We present a comprehensive review of 46 methods for PGS construction. We connect the majority of these methods through a multiple linear regression framework which can be instrumental for understanding their prediction performance for traits with distinct genetic architectures. We discuss the practical considerations of PGS analysis as well as challenges and future directions of PGS method development. We hope our review serves as a useful reference both for statistical geneticists who develop PGS methods and for data analysts who perform PGS analysis.
Keywords: complex traits; genetic prediction; genome-wide association studies; polygenic risk scores; polygenic scores; statistical methods.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests No interests are declared.
Figures
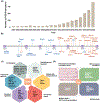

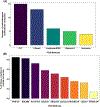

References
-
- Andersson L and Georges M (2004) Domestic-animal genomics: deciphering the genetics of complex traits. Nat. Rev. Genet 5, 202–212 - PubMed
-
- Frazer KA et al. (2009) Human genetic variation and its contribution to complex traits. Nat. Rev. Genet 10, 241–251 - PubMed
-
- McCarthy MI et al. (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet 9, 356–369 - PubMed
-
- Martincorena I and Campbell PJ (2015) Somatic mutation in cancer and normal cells. Science 349, 1483–1489 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

