Mitochondrial compartmentalization: emerging themes in structure and function
- PMID: 34244035
- PMCID: PMC11008732
- DOI: 10.1016/j.tibs.2021.06.003
Mitochondrial compartmentalization: emerging themes in structure and function
Abstract
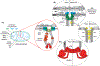
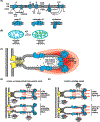
Within cellular structures, compartmentalization is the concept of spatial segregation of macromolecules, metabolites, and biochemical pathways. Therefore, this concept bridges organellar structure and function. Mitochondria are morphologically complex, partitioned into several subcompartments by a topologically elaborate two-membrane system. They are also dynamically polymorphic, undergoing morphogenesis events with an extent and frequency that is only now being appreciated. Thus, mitochondrial compartmentalization is something that must be considered both spatially and temporally. Here, we review new developments in how mitochondrial structure is established and regulated, the factors that underpin the distribution of lipids and proteins, and how they spatially demarcate locations of myriad mitochondrial processes. Consistent with its pre-eminence, disturbed mitochondrial compartmentalization contributes to the dysfunction associated with heritable and aging-related diseases.
Keywords: bioenergetics; cristae; macromolecular trafficking; mitochondria; morphogenesis; ultrastructure.
Published by Elsevier Ltd.
Conflict of interest statement
Declaration of interests None declared by authors.
Figures

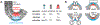
References
-
- Cogliati S. et al. (2016) Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem Sci 41 (3), 261–273. - PubMed
-
- Quintana-Cabrera R. et al. (2018) Who and how in the regulation of mitochondrial cristae shape and function. Biochem Biophys Res Commun 500 (1), 94–101. - PubMed
-
- Giacomello M. et al. (2020) The cell biology of mitochondrial membrane dynamics. Nat Rev Mol Cell Biol 21 (4), 204–224. - PubMed
-
- Li W. et al. (2020) Dynamic organization of intracellular organelle networks. Wiley Interdiscip Rev Syst Biol Med, e1505. - PubMed
-
- Rasmussen N. (1995) Mitochondrial structure and the practice of cell biology in the 1950s. J Hist Biol 28 (3), 381–429. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

