Multiplexed imaging analysis of the tumor-immune microenvironment reveals predictors of outcome in triple-negative breast cancer
- PMID: 34244605
- PMCID: PMC8271023
- DOI: 10.1038/s42003-021-02361-1
Multiplexed imaging analysis of the tumor-immune microenvironment reveals predictors of outcome in triple-negative breast cancer
Abstract
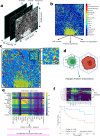
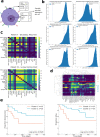
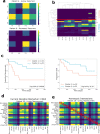
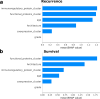
Triple-negative breast cancer, the poorest-prognosis breast cancer subtype, lacks clinically approved biomarkers for patient risk stratification and treatment management. Prior literature has shown that interrogation of the tumor-immune microenvironment may be a promising approach to fill these gaps. Recently developed high-dimensional tissue imaging technology, such as multiplexed ion beam imaging, provide spatial context to protein expression in the microenvironment, allowing in-depth characterization of cellular processes. We demonstrate that profiling the functional proteins involved in cell-to-cell interactions in the microenvironment can predict recurrence and overall survival. We highlight the immunological relevance of the immunoregulatory proteins PD-1, PD-L1, IDO, and Lag3 by tying interactions involving them to recurrence and survival. Multivariate analysis reveals that our methods provide additional prognostic information compared to clinical variables. In this work, we present a computational pipeline for the examination of the tumor-immune microenvironment using multiplexed ion beam imaging that produces interpretable results, and is generalizable to other cancer types.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

