Structure and function of retroviral integrase
- PMID: 34244677
- PMCID: PMC8671357
- DOI: 10.1038/s41579-021-00586-9
Structure and function of retroviral integrase
Abstract
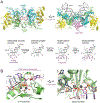
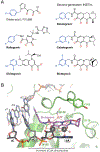
A hallmark of retroviral replication is establishment of the proviral state, wherein a DNA copy of the viral RNA genome is stably incorporated into a host cell chromosome. Integrase is the viral enzyme responsible for the catalytic steps involved in this process, and integrase strand transfer inhibitors are widely used to treat people living with HIV. Over the past decade, a series of X-ray crystallography and cryogenic electron microscopy studies have revealed the structural basis of retroviral DNA integration. A variable number of integrase molecules congregate on viral DNA ends to assemble a conserved intasome core machine that facilitates integration. The structures additionally informed on the modes of integrase inhibitor action and the means by which HIV acquires drug resistance. Recent years have witnessed the development of allosteric integrase inhibitors, a highly promising class of small molecules that antagonize viral morphogenesis. In this Review, we explore recent insights into the organization and mechanism of the retroviral integration machinery and highlight open questions as well as new directions in the field.
© 2021. Springer Nature Limited.
Conflict of interest statement
Conflicts of Interest
A.N.E. consults for ViiV Healthcare, Co. The other authors have no conflicts to declare.
Figures
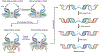
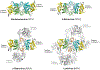
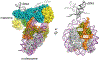

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

