Anti-tumor effects of RTX-240: an engineered red blood cell expressing 4-1BB ligand and interleukin-15
- PMID: 34244816
- PMCID: PMC8360899
- DOI: 10.1007/s00262-021-03001-7
Anti-tumor effects of RTX-240: an engineered red blood cell expressing 4-1BB ligand and interleukin-15
Abstract
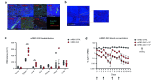
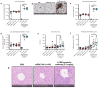
Recombinant agonists that activate co-stimulatory and cytokine receptors have shown limited clinical anticancer utility, potentially due to narrow therapeutic windows, the need for coordinated activation of co-stimulatory and cytokine pathways and the failure of agonistic antibodies to recapitulate signaling by endogenous ligands. RTX-240 is a genetically engineered red blood cell expressing 4-1BBL and IL-15/IL-15Rα fusion (IL-15TP). RTX-240 is designed to potently and simultaneously stimulate the 4-1BB and IL-15 pathways, thereby activating and expanding T cells and NK cells, while potentially offering an improved safety profile through restricted biodistribution. We assessed the ability of RTX-240 to expand and activate T cells and NK cells and evaluated the in vivo efficacy, pharmacodynamics and tolerability using murine models. Treatment of PBMCs with RTX-240 induced T cell and NK cell activation and proliferation. In vivo studies using mRBC-240, a mouse surrogate for RTX-240, revealed biodistribution predominantly to the red pulp of the spleen, leading to CD8 + T cell and NK cell expansion. mRBC-240 was efficacious in a B16-F10 melanoma model and led to increased NK cell infiltration into the lungs. mRBC-240 significantly inhibited CT26 tumor growth, in association with an increase in tumor-infiltrating proliferating and cytotoxic CD8 + T cells. mRBC-240 was tolerated and showed no evidence of hepatic injury at the highest feasible dose, compared with a 4-1BB agonistic antibody. RTX-240 promotes T cell and NK cell activity in preclinical models and shows efficacy and an improved safety profile. Based on these data, RTX-240 is now being evaluated in a clinical trial.
Keywords: CD8-Positive T-Lymphocytes; Cell engineering; Immunotherapy; Investigational therapies; Natural killer cells.
© 2021. The Author(s).
Figures








References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

