The transcription factor code in iPSC reprogramming
- PMID: 34246082
- PMCID: PMC9469655
- DOI: 10.1016/j.gde.2021.06.003
The transcription factor code in iPSC reprogramming
Abstract
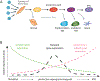
Transcription factor (TF)-induced reprogramming of somatic cells across lineages and to induced pluripotent stem cells (iPSCs) has revealed a remarkable plasticity of differentiated cells and presents great opportunities for generating clinically relevant cell types for disease modeling and regenerative medicine. The understanding of iPSC reprogramming provides insights into the mechanisms that safeguard somatic cell identity, drive epigenetic reprogramming, and underlie cell fate specification in vivo. The combinatorial action of TFs has emerged as the key mechanism for the direct and indirect effects of reprogramming factors that induce the remodelling of the enhancer landscape. The interplay of TFs in iPSC reprogramming also yields trophectoderm- and extraembryonic endoderm-like cell populations, uncovering an intriguing plasticity of cell states and opening new avenues for exploring cell fate decisions during early embryogenesis.
Copyright © 2021 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Interest
The authors declare no conflict of interest.
Figures

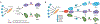
References
-
- Spitz F, Furlong EE: Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 2012, 13:613–626. - PubMed
-
- Takahashi K, Yamanaka S: Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126:663–676. - PubMed
-
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al.: Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 2007, 1:55–70. - PubMed
-
- Nefzger CM, Rossello FJ, Chen J, Liu X, Knaupp AS, Firas J, Paynter JM, Pflueger J, Buckberry S, Lim SM, et al.: Cell Type of Origin Dictates the Route to Pluripotency. Cell Rep 2017, 21:2649–2660. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

