In Silico Exploration of Phytoconstituents From Phyllanthus emblica and Aegle marmelos as Potential Therapeutics Against SARS-CoV-2 RdRp
- PMID: 34248355
- PMCID: PMC8236766
- DOI: 10.1177/11779322211027403
In Silico Exploration of Phytoconstituents From Phyllanthus emblica and Aegle marmelos as Potential Therapeutics Against SARS-CoV-2 RdRp
Abstract
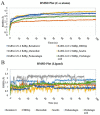
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) worldwide has increased the importance of computational tools to design a drug or vaccine in reduced time with minimum risk. Earlier studies have emphasized the important role of RNA-dependent RNA polymerase (RdRp) in SARS-CoV-2 replication as a potential drug target. In our study, comprehensive computational approaches were applied to identify potential compounds targeting RdRp of SARS-CoV-2. To study the binding affinity and stability of the phytocompounds from Phyllanthus emblica and Aegel marmelos within the defined binding site of SARS-CoV-2 RdRp, they were subjected to molecular docking, 100 ns molecular dynamics (MD) simulation followed by post-simulation analysis. Furthermore, to assess the importance of features involved in the strong binding affinity, molecular field-based similarity analysis was performed. Based on comparative molecular docking and simulation studies of the selected phytocompounds with SARS-CoV-2 RdRp revealed that EBDGp possesses a stronger binding affinity (-23.32 kcal/mol) and stability than other phytocompounds and reference compound, Remdesivir (-19.36 kcal/mol). Molecular field-based similarity profiling has supported our study in the validation of the importance of the presence of hydroxyl groups in EBDGp, involved in increasing its binding affinity toward SARS-CoV-2 RdRp. Molecular docking and dynamic simulation results confirmed that EBDGp has better inhibitory potential than Remdesivir and can be an effective novel drug for SARS-CoV-2 RdRp. Furthermore, binding free energy calculations confirmed the higher stability of the SARS-CoV-2 RdRp-EBDGp complex. These results suggest that the EBDGp compound may emerge as a promising drug against SARS-CoV-2 and hence requires further experimental validation.
Keywords: Aegle marmelos SARS-CoV-2 RdRp; MD simulation; Phyllanthus emblica; drug repurposing; molecular docking.
© The Author(s) 2021.
Conflict of interest statement
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures









Similar articles
-
Revealing the Inhibition Mechanism of RNA-Dependent RNA Polymerase (RdRp) of SARS-CoV-2 by Remdesivir and Nucleotide Analogues: A Molecular Dynamics Simulation Study.J Phys Chem B. 2020 Nov 25;124(47):10641-10652. doi: 10.1021/acs.jpcb.0c06747. Epub 2020 Nov 15. J Phys Chem B. 2020. PMID: 33190493
-
Identification of SARS-CoV-2 RNA dependent RNA polymerase inhibitors using pharmacophore modelling, molecular docking and molecular dynamics simulation approaches.J Biomol Struct Dyn. 2022;40(24):13366-13377. doi: 10.1080/07391102.2021.1987329. Epub 2021 Oct 12. J Biomol Struct Dyn. 2022. PMID: 34637693
-
Integrated in Silico and in Vitro Studies of Rutin's Potential against SARS-CoV-2 through the Inhibition of the RNA-dependent RNA Polymerase.Curr Med Chem. 2025 Jan 2. doi: 10.2174/0109298673339634241210151734. Online ahead of print. Curr Med Chem. 2025. PMID: 39757685
-
State-of-the-Art Molecular Dynamics Simulation Studies of RNA-Dependent RNA Polymerase of SARS-CoV-2.Int J Mol Sci. 2022 Sep 8;23(18):10358. doi: 10.3390/ijms231810358. Int J Mol Sci. 2022. PMID: 36142270 Free PMC article. Review.
-
Repurposing potential of Ayurvedic medicinal plants derived active principles against SARS-CoV-2 associated target proteins revealed by molecular docking, molecular dynamics and MM-PBSA studies.Biomed Pharmacother. 2021 May;137:111356. doi: 10.1016/j.biopha.2021.111356. Epub 2021 Feb 3. Biomed Pharmacother. 2021. PMID: 33561649 Free PMC article.
Cited by
-
Effect of polyphenols against complications of COVID-19: current evidence and potential efficacy.Pharmacol Rep. 2024 Apr;76(2):307-327. doi: 10.1007/s43440-024-00585-6. Epub 2024 Mar 18. Pharmacol Rep. 2024. PMID: 38498260 Review.
-
A hypothesis on designing strategy of effective RdRp inhibitors for the treatment of SARS-CoV-2.3 Biotech. 2023 Jan;13(1):12. doi: 10.1007/s13205-022-03430-w. Epub 2022 Dec 16. 3 Biotech. 2023. PMID: 36532857 Free PMC article. Review.
-
Exploration of phytochemical compounds against Marburg virus using QSAR, molecular dynamics, and free energy landscape.Mol Divers. 2024 Oct;28(5):3261-3278. doi: 10.1007/s11030-023-10753-0. Epub 2023 Nov 5. Mol Divers. 2024. PMID: 37925643
-
In silico Screening of Potential SARS-CoV-2 Main Protease Inhibitors from Thymus schimperi.Adv Appl Bioinform Chem. 2023 Jan 18;16:1-13. doi: 10.2147/AABC.S393084. eCollection 2023. Adv Appl Bioinform Chem. 2023. PMID: 36699952 Free PMC article.
-
Genome of Phyllanthus emblica: the medicinal plant Amla with super antioxidant properties.Front Plant Sci. 2023 Sep 1;14:1210078. doi: 10.3389/fpls.2023.1210078. eCollection 2023. Front Plant Sci. 2023. PMID: 37727852 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

