A Master Regulator BrpR Coordinates the Expression of Multiple Loci for Robust Biofilm and Rugose Colony Development in Vibrio vulnificus
- PMID: 34248894
- PMCID: PMC8268162
- DOI: 10.3389/fmicb.2021.679854
A Master Regulator BrpR Coordinates the Expression of Multiple Loci for Robust Biofilm and Rugose Colony Development in Vibrio vulnificus
Abstract
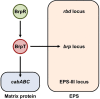
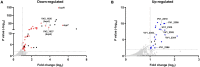
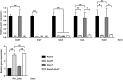
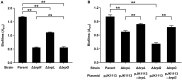
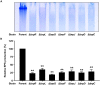
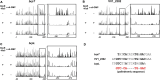
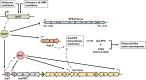
Vibrio vulnificus, a fulminating human pathogen, forms biofilms to enhance its survival in nature and pathogenicity during host infection. BrpR is the transcriptional regulator governing robust biofilm and rugose colony formation in V. vulnificus, but little is known about both the direct regulon of BrpR and the role of BrpR in regulation of downstream genes. In this study, transcript analyses revealed that BrpR is highly expressed and thus strongly regulates the downstream gene in the stationary and elevated cyclic di-GMP conditions. Transcriptome analyses discovered the genes, whose expression is affected by BrpR but not by the downstream regulator BrpT. Two unnamed adjacent genes (VV2_1626-1627) were newly identified among the BrpR regulon and designated as brpL and brpG in this study. Genetic analyses showed that the deletion of brpL and brpG impairs the biofilm and rugose colony formation, indicating that brpLG plays a crucial role in the development of BrpR-regulated biofilm phenotypes. Comparison of the colony morphology and exopolysaccharide (EPS) production suggested that although the genetic location and regulation of brpLG are distinct from the brp locus, brpABCDFHIJK (VV2_1574-1582), brpLG is also responsible for the robust EPS production together with the brp locus genes. Electrophoretic mobility shift assays and DNase I protection assays demonstrated that BrpR regulates the expression of downstream genes in distinct loci by directly binding to their upstream regions, revealing a palindromic binding sequence. Altogether, this study suggests that BrpR is a master regulator coordinating the expression of multiple loci responsible for EPS production and thus, contributing to the robust biofilm and rugose colony formation of V. vulnificus.
Keywords: Vibrio vulnificus; biofilm; colony morphology; exopolysaccharide; transcriptional regulator.
Copyright © 2021 Hwang, Im and Choi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Two novel genes identified by large-scale transcriptomic analysis are essential for biofilm and rugose colony development of Vibrio vulnificus.PLoS Pathog. 2023 Jan 19;19(1):e1011064. doi: 10.1371/journal.ppat.1011064. eCollection 2023 Jan. PLoS Pathog. 2023. PMID: 36656902 Free PMC article.
-
Complex Control of a Genomic Island Governing Biofilm and Rugose Colony Development in Vibrio vulnificus.J Bacteriol. 2018 Jul 25;200(16):e00190-18. doi: 10.1128/JB.00190-18. Print 2018 Aug 15. J Bacteriol. 2018. PMID: 29760209 Free PMC article.
-
A Regulatory Network Controls cabABC Expression Leading to Biofilm and Rugose Colony Development in Vibrio vulnificus.Front Microbiol. 2020 Jan 17;10:3063. doi: 10.3389/fmicb.2019.03063. eCollection 2019. Front Microbiol. 2020. PMID: 32010109 Free PMC article.
-
Identification and characterization of a small molecule BFstatin inhibiting BrpR, the transcriptional regulator for biofilm formation of Vibrio vulnificus.Front Microbiol. 2024 Sep 9;15:1468567. doi: 10.3389/fmicb.2024.1468567. eCollection 2024. Front Microbiol. 2024. PMID: 39314881 Free PMC article.
-
Identification of a c-di-GMP-regulated polysaccharide locus governing stress resistance and biofilm and rugose colony formation in Vibrio vulnificus.Infect Immun. 2010 Mar;78(3):1390-402. doi: 10.1128/IAI.01188-09. Epub 2010 Jan 11. Infect Immun. 2010. PMID: 20065022 Free PMC article.
Cited by
-
Genome-wide phenotypic profiling of transcription factors and identification of novel targets to control the virulence of Vibrio vulnificus.Nucleic Acids Res. 2025 Jan 24;53(3):gkae1238. doi: 10.1093/nar/gkae1238. Nucleic Acids Res. 2025. PMID: 39704106 Free PMC article.
-
Two novel genes identified by large-scale transcriptomic analysis are essential for biofilm and rugose colony development of Vibrio vulnificus.PLoS Pathog. 2023 Jan 19;19(1):e1011064. doi: 10.1371/journal.ppat.1011064. eCollection 2023 Jan. PLoS Pathog. 2023. PMID: 36656902 Free PMC article.
-
Transition of Dephospho-DctD to the Transcriptionally Active State via Interaction with Dephospho-IIAGlc.mBio. 2022 Apr 26;13(2):e0383921. doi: 10.1128/mbio.03839-21. Epub 2022 Mar 21. mBio. 2022. PMID: 35311533 Free PMC article.
-
Spatio-genetically coordinated TPR domain-containing proteins modulate c-di-GMP signaling in Vibrio vulnificus.PLoS Pathog. 2025 Jul 16;21(7):e1013353. doi: 10.1371/journal.ppat.1013353. eCollection 2025 Jul. PLoS Pathog. 2025. PMID: 40668869 Free PMC article.
-
CarRS Two-Component System Essential for Polymyxin B Resistance of Vibrio vulnificus Responds to Multiple Host Environmental Signals.Microbiol Spectr. 2023 Aug 17;11(4):e0030523. doi: 10.1128/spectrum.00305-23. Epub 2023 Jun 8. Microbiol Spectr. 2023. PMID: 37289068 Free PMC article.
References
-
- Bechet E., Gruszczyk J., Terreux R., Gueguen-Chaignon V., Vigouroux A., Obadia B., et al. (2010). Identification of structural and molecular determinants of the tyrosine-kinase Wzc and implications in capsular polysaccharide export. Mol. Microbiol. 77 1315–1325. 10.1111/j.1365-2958.2010.07291.x - DOI - PubMed
LinkOut - more resources
Full Text Sources

