Oral Microbiome Dysbiosis Is Associated With Symptoms Severity and Local Immune/Inflammatory Response in COVID-19 Patients: A Cross-Sectional Study
- PMID: 34248910
- PMCID: PMC8261071
- DOI: 10.3389/fmicb.2021.687513
Oral Microbiome Dysbiosis Is Associated With Symptoms Severity and Local Immune/Inflammatory Response in COVID-19 Patients: A Cross-Sectional Study
Abstract
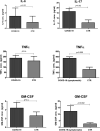
The human oral microbiome (HOM) is the second largest microbial community after the gut and can impact the onset and progression of several localized and systemic diseases, including those of viral origin, especially for viruses entering the body via the oropharynx. However, this important aspect has not been clarified for the new pandemic human coronavirus SARS-CoV-2, causing COVID-19 disease, despite it being one of the many respiratory viruses having the oropharynx as the primary site of replication. In particular, no data are available about the non-bacterial components of the HOM (fungi, viruses), which instead has been shown to be crucial for other diseases. Consistent with this, this study aimed to define the HOM in COVID-19 patients, to evidence any association between its profile and the clinical disease. Seventy-five oral rinse samples were analyzed by Whole Genome Sequencing (WGS) to simultaneously identify oral bacteria, fungi, and viruses. To correlate the HOM profile with local virus replication, the SARS-CoV-2 amount in the oral cavity was quantified by digital droplet PCR. Moreover, local inflammation and secretory immune response were also assessed, respectively by measuring the local release of pro-inflammatory cytokines (L-6, IL-17, TNFα, and GM-CSF) and the production of secretory immunoglobulins A (sIgA). The results showed the presence of oral dysbiosis in COVID-19 patients compared to matched controls, with significantly decreased alpha-diversity value and lower species richness in COVID-19 subjects. Notably, oral dysbiosis correlated with symptom severity (p = 0.006), and increased local inflammation (p < 0.01). In parallel, a decreased mucosal sIgA response was observed in more severely symptomatic patients (p = 0.02), suggesting that local immune response is important in the early control of virus infection and that its correct development is influenced by the HOM profile. In conclusion, the data presented here suggest that the HOM profile may be important in defining the individual susceptibility to SARS-CoV-2 infection, facilitating inflammation and virus replication, or rather, inducing a protective IgA response. Although it is not possible to determine whether the alteration in the microbial community is the cause or effect of the SARS-CoV-2 replication, these parameters may be considered as markers for personalized therapy and vaccine development.
Keywords: COVID-19; inflammatory cytokines; oral microbiome; secretory IgA; symptom severity.
Copyright © 2021 Soffritti, D’Accolti, Fabbri, Passaro, Manfredini, Zuliani, Libanore, Franchi, Contini and Caselli.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Andrews T., Thompson M., Buckley D. I., Heneghan C., Deyo R., Redmond N., et al. (2012). Interventions to influence consulting and antibiotic use for acute respiratory tract infections in children: a systematic review and meta-analysis. PLoS One 7:e30334. 10.1371/journal.pone.0030334 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

